Wastewater-based prediction of COVID-19 cases using a highly sensitive SARS-CoV-2 RNA detection method combined with mathematical modeling
- PMID: 36867995
- PMCID: PMC9824953
- DOI: 10.1016/j.envint.2023.107743
Wastewater-based prediction of COVID-19 cases using a highly sensitive SARS-CoV-2 RNA detection method combined with mathematical modeling
Abstract
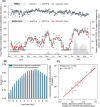
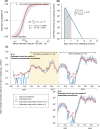
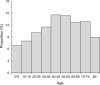
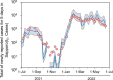
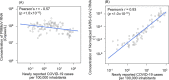
Wastewater-based epidemiology (WBE) has the potential to predict COVID-19 cases; however, reliable methods for tracking SARS-CoV-2 RNA concentrations (CRNA) in wastewater are lacking. In the present study, we developed a highly sensitive method (EPISENS-M) employing adsorption-extraction, followed by one-step RT-Preamp and qPCR. The EPISENS-M allowed SARS-CoV-2 RNA detection from wastewater at 50 % detection rate when newly reported COVID-19 cases exceed 0.69/100,000 inhabitants in a sewer catchment. Using the EPISENS-M, a longitudinal WBE study was conducted between 28 May 2020 and 16 June 2022 in Sapporo City, Japan, revealing a strong correlation (Pearson's r = 0.94) between CRNA and the newly COVID-19 cases reported by intensive clinical surveillance. Based on this dataset, a mathematical model was developed based on viral shedding dynamics to estimate the newly reported cases using CRNA data and recent clinical data prior to sampling day. This developed model succeeded in predicting the cumulative number of newly reported cases after 5 days of sampling day within a factor of √2 and 2 with a precision of 36 % (16/44) and 64 % (28/44), respectively. By applying this model framework, another estimation mode was developed without the recent clinical data, which successfully predicted the number of COVID-19 cases for the succeeding 5 days within a factor of √2 and 2 with a precision of 39 % (17/44) and 66 % (29/44), respectively. These results demonstrated that the EPISENS-M method combined with the mathematical model can be a powerful tool for predicting COVID-19 cases, especially in the absence of intensive clinical surveillance.
Keywords: COVID-19; EPISENS-M; Mathematical model; Quantification method; SARS-CoV-2; Wastewater-based epidemiology.
Copyright © 2023. Published by Elsevier Ltd.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Ahmed, W. et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Science of the Total Environment vol. 805 Preprint at https://doi.org/10.1016/j.scitotenv.2021.149877 (2022). - PMC - PubMed
Further reading
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

