hPSC-derived sacral neural crest enables rescue in a severe model of Hirschsprung's disease
- PMID: 36868194
- PMCID: PMC10034921
- DOI: 10.1016/j.stem.2023.02.003
hPSC-derived sacral neural crest enables rescue in a severe model of Hirschsprung's disease
Abstract
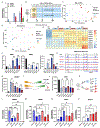
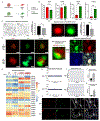
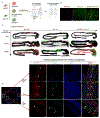
The enteric nervous system (ENS) is derived from both the vagal and sacral component of the neural crest (NC). Here, we present the derivation of sacral ENS precursors from human PSCs via timed exposure to FGF, WNT, and GDF11, which enables posterior patterning and transition from posterior trunk to sacral NC identity, respectively. Using a SOX2::H2B-tdTomato/T::H2B-GFP dual reporter hPSC line, we demonstrate that both trunk and sacral NC emerge from a double-positive neuro-mesodermal progenitor (NMP). Vagal and sacral NC precursors yield distinct neuronal subtypes and migratory behaviors in vitro and in vivo. Remarkably, xenografting of both vagal and sacral NC lineages is required to rescue a mouse model of total aganglionosis, suggesting opportunities in the treatment of severe forms of Hirschsprung's disease.
Keywords: GDF11; Hirschsprung's disease; axial patterning; axial progenitors; cell therapy; directed differentiation; enteric nervous system; nervous system disorder; neural crest development; pluripotent stem cells; regenerative medicine; sacral neural crest.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests L.S. is a scientific co-founder and consultant and has received sponsored research support for work related to this study from Bluerock Therapeutics. L.S. and Y.F. are inventors of a patent application filed by Memorial Sloan Kettering Cancer Center on the methods described in this study.
Figures




References
-
- Gershon M (1999). The second brain: a groundbreaking new understanding of nervous disorders of the stomach and intestine (HarperCollins; ).
-
- Bodian M, and Carter OO (1963). A family study of Hirschsprung’s disease. Annals of Human Genetics 26, 261–277.
-
- Bolande RP (1974). The neurocristopathies: a unifying concept of disease arising in neural crest maldevelopment. Human pathology 5, 409–429. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

