Apoptotic cell fragments locally activate tingible body macrophages in the germinal center
- PMID: 36868219
- PMCID: PMC7614509
- DOI: 10.1016/j.cell.2023.02.004
Apoptotic cell fragments locally activate tingible body macrophages in the germinal center
Abstract
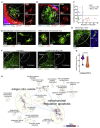
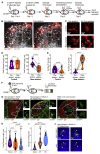
Germinal centers (GCs) that form within lymphoid follicles during antibody responses are sites of massive cell death. Tingible body macrophages (TBMs) are tasked with apoptotic cell clearance to prevent secondary necrosis and autoimmune activation by intracellular self antigens. We show by multiple redundant and complementary methods that TBMs derive from a lymph node-resident, CD169-lineage, CSF1R-blockade-resistant precursor that is prepositioned in the follicle. Non-migratory TBMs use cytoplasmic processes to chase and capture migrating dead cell fragments using a "lazy" search strategy. Follicular macrophages activated by the presence of nearby apoptotic cells can mature into TBMs in the absence of GCs. Single-cell transcriptomics identified a TBM cell cluster in immunized lymph nodes which upregulated genes involved in apoptotic cell clearance. Thus, apoptotic B cells in early GCs trigger activation and maturation of follicular macrophages into classical TBMs to clear apoptotic debris and prevent antibody-mediated autoimmune diseases.
Keywords: B cells; apoptosis; autoimmunity; germinal center; tingible body macrophages.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Comment in
-
Tingible body macrophages: Gargantuan chameleons of the germinal center.J Exp Med. 2023 Apr 3;220(4):e20230250. doi: 10.1084/jem.20230250. Epub 2023 Mar 14. J Exp Med. 2023. PMID: 36917028 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

