Mimicking Tumor Cell Heterogeneity of Colorectal Cancer in a Patient-derived Organoid-Fibroblast Model
- PMID: 36868311
- PMCID: PMC10141529
- DOI: 10.1016/j.jcmgh.2023.02.014
Mimicking Tumor Cell Heterogeneity of Colorectal Cancer in a Patient-derived Organoid-Fibroblast Model
Abstract
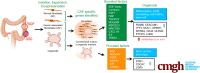
Background & aims: Patient-derived organoid cancer models are generated from epithelial tumor cells and reflect tumor characteristics. However, they lack the complexity of the tumor microenvironment, which is a key driver of tumorigenesis and therapy response. Here, we developed a colorectal cancer organoid model that incorporates matched epithelial cells and stromal fibroblasts.
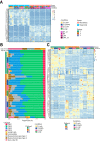
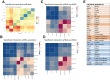
Methods: Primary fibroblasts and tumor cells were isolated from colorectal cancer specimens. Fibroblasts were characterized for their proteome, secretome, and gene expression signatures. Fibroblast/organoid co-cultures were analyzed by immunohistochemistry and compared with their tissue of origin, as well as on gene expression levels compared with standard organoid models. Bioinformatics deconvolution was used to calculate cellular proportions of cell subsets in organoids based on single-cell RNA sequencing data.
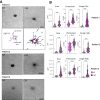
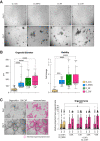
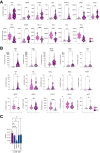
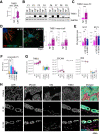
Results: Normal primary fibroblasts, isolated from tumor adjacent tissue, and cancer associated fibroblasts retained their molecular characteristics in vitro, including higher motility of cancer associated compared with normal fibroblasts. Importantly, both cancer-associated fibroblasts and normal fibroblasts supported cancer cell proliferation in 3D co-cultures, without the addition of classical niche factors. Organoids grown together with fibroblasts displayed a larger cellular heterogeneity of tumor cells compared with mono-cultures and closely resembled the in vivo tumor morphology. Additionally, we observed a mutual crosstalk between tumor cells and fibroblasts in the co-cultures. This was manifested by considerably deregulated pathways such as cell-cell communication and extracellular matrix remodeling in the organoids. Thrombospondin-1 was identified as a critical factor for fibroblast invasiveness.
Conclusion: We developed a physiological tumor/stroma model, which will be vital as a personalized tumor model to study disease mechanisms and therapy response in colorectal cancer.
Keywords: Cancer; Co-cultures; Colorectal Cancer; Fibroblasts; Organoids.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Arnold M., Sierra M.S., Laversanne M., et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. - PubMed
-
- Ooft S.N., Weeber F., Dijkstra K.K., et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019;11 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

