Perspective: The current state of Cre driver mouse lines in skeletal research: Challenges and opportunities
- PMID: 36868507
- PMCID: PMC10087282
- DOI: 10.1016/j.bone.2023.116719
Perspective: The current state of Cre driver mouse lines in skeletal research: Challenges and opportunities
Abstract
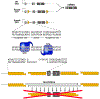
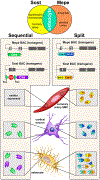
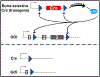
The Cre/Lox system has revolutionized the ability of biomedical researchers to ask very specific questions about the function of individual genes in specific cell types at specific times during development and/or disease progression in a variety of animal models. This is true in the skeletal biology field, and numerous Cre driver lines have been created to foster conditional gene manipulation in specific subpopulations of bone cells. However, as our ability to scrutinize these models increases, an increasing number of issues have been identified with most driver lines. All existing skeletal Cre mouse models exhibit problems in one or more of the following three areas: (1) cell type specificity-avoiding Cre expression in unintended cell types; (2) Cre inducibility-improving the dynamic range for Cre in inducible models (negligible Cre activity before induction and high Cre activity after induction); and (3) Cre toxicity-reducing the unwanted biological effects of Cre (beyond loxP recombination) on cellular processes and tissue health. These issues are hampering progress in understanding the biology of skeletal disease and aging, and consequently, identification of reliable therapeutic opportunities. Skeletal Cre models have not advanced technologically in decades despite the availability of improved tools, including multi-promoter-driven expression of permissive or fragmented recombinases, new dimerization systems, and alternative forms of recombinases and DNA sequence targets. We review the current state of skeletal Cre driver lines, and highlight some of the successes, failures, and opportunities to improve fidelity in the skeleton, based on successes pioneered in other areas of biomedical science.
Keywords: Cre; Floxed allele; LoxP; Recombination; Skeletal models.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest All authors have declared that no conflicts of interest exist. Financial support was provided by the NIH (AR053237 to AGR; AG069489 to RBC) and the US Department of Veterans Affairs (BX001478, IK6 BX003783, and I01 BX005861 to AGR).
Figures
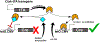


References
-
- Sakharkar MK, Chow VT & Kangueane P (2004) Distributions of exons and introns in the human genome. In Silico Biol 4(4):387–393 - PubMed
-
- Gu H, Marth JD, Orban PC, Mossmann H & Rajewsky K (1994) Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science 265(5168):103–106 - PubMed
-
- Feil R, Wagner J, Metzger D & Chambon P (1997) Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 237(3):752–757 - PubMed
-
- Couasnay G, Madel MB, Lim J, Lee B & Elefteriou F (2021) Sites of Cre-recombinase activity in mouse lines targeting skeletal cells. J Bone Miner Res 36(9):1661–1679 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

