An autoimmune pleiotropic SNP modulates IRF5 alternative promoter usage through ZBTB3-mediated chromatin looping
- PMID: 36869052
- PMCID: PMC9984425
- DOI: 10.1038/s41467-023-36897-z
An autoimmune pleiotropic SNP modulates IRF5 alternative promoter usage through ZBTB3-mediated chromatin looping
Abstract
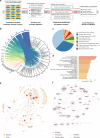
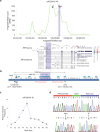
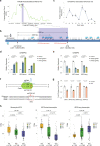
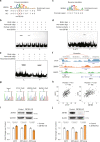
Genetic sharing is extensively observed for autoimmune diseases, but the causal variants and their underlying molecular mechanisms remain largely unknown. Through systematic investigation of autoimmune disease pleiotropic loci, we found most of these shared genetic effects are transmitted from regulatory code. We used an evidence-based strategy to functionally prioritize causal pleiotropic variants and identify their target genes. A top-ranked pleiotropic variant, rs4728142, yielded many lines of evidence as being causal. Mechanistically, the rs4728142-containing region interacts with the IRF5 alternative promoter in an allele-specific manner and orchestrates its upstream enhancer to regulate IRF5 alternative promoter usage through chromatin looping. A putative structural regulator, ZBTB3, mediates the allele-specific loop to promote IRF5-short transcript expression at the rs4728142 risk allele, resulting in IRF5 overactivation and M1 macrophage polarization. Together, our findings establish a causal mechanism between the regulatory variant and fine-scale molecular phenotype underlying the dysfunction of pleiotropic genes in human autoimmunity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Allele-specific chromatin looping promotes IRF5-dependent autoimmunity.Nat Rev Rheumatol. 2023 May;19(5):256. doi: 10.1038/s41584-023-00956-y. Nat Rev Rheumatol. 2023. PMID: 37012360 No abstract available.
Similar articles
-
Integrative Functional Genomics Identifies Systemic Lupus Erythematosus Causal Genetic Variant in the IRF5 Risk Locus.Arthritis Rheumatol. 2023 Apr;75(4):574-585. doi: 10.1002/art.42390. Epub 2023 Feb 8. Arthritis Rheumatol. 2023. PMID: 36245280
-
An Allele-Specific Functional SNP Associated with Two Systemic Autoimmune Diseases Modulates IRF5 Expression by Long-Range Chromatin Loop Formation.J Invest Dermatol. 2020 Feb;140(2):348-360.e11. doi: 10.1016/j.jid.2019.06.147. Epub 2019 Aug 15. J Invest Dermatol. 2020. PMID: 31421124
-
The IRF5-TNPO3 association with systemic lupus erythematosus has two components that other autoimmune disorders variably share.Hum Mol Genet. 2015 Jan 15;24(2):582-96. doi: 10.1093/hmg/ddu455. Epub 2014 Sep 8. Hum Mol Genet. 2015. PMID: 25205108 Free PMC article.
-
Interferon regulatory factor 5 in the pathogenesis of systemic lupus erythematosus.Clin Dev Immunol. 2012;2012:780436. doi: 10.1155/2012/780436. Epub 2012 Nov 1. Clin Dev Immunol. 2012. PMID: 23251221 Free PMC article. Review.
-
IRF5-mediated immune responses and its implications in immunological disorders.Int Rev Immunol. 2018;37(5):229-248. doi: 10.1080/08830185.2018.1469629. Epub 2018 Jul 9. Int Rev Immunol. 2018. PMID: 29985675 Review.
Cited by
-
Integrative high-throughput enhancer surveying and functional verification divulges a YY2-condensed regulatory axis conferring risk for osteoporosis.Cell Genom. 2024 Mar 13;4(3):100501. doi: 10.1016/j.xgen.2024.100501. Epub 2024 Feb 8. Cell Genom. 2024. PMID: 38335956 Free PMC article.
-
Identification and characterization of whole blood gene expression and splicing quantitative trait loci during early to mid-lactation of dairy cattle.BMC Genomics. 2024 May 6;25(1):445. doi: 10.1186/s12864-024-10346-7. BMC Genomics. 2024. PMID: 38711039 Free PMC article.
-
3D genome organization and epigenetic regulation in autoimmune diseases.Front Immunol. 2023 Jun 6;14:1196123. doi: 10.3389/fimmu.2023.1196123. eCollection 2023. Front Immunol. 2023. PMID: 37346038 Free PMC article. Review.
-
Allele-specific chromatin looping promotes IRF5-dependent autoimmunity.Nat Rev Rheumatol. 2023 May;19(5):256. doi: 10.1038/s41584-023-00956-y. Nat Rev Rheumatol. 2023. PMID: 37012360 No abstract available.
-
Analysis of biased allelic enhancer activity of schizophrenia-linked common variants.Commun Biol. 2025 Jul 10;8(1):1034. doi: 10.1038/s42003-025-08456-3. Commun Biol. 2025. PMID: 40640362 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

