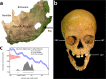
Rickettsia felis DNA recovered from a child who lived in southern Africa 2000 years ago
- PMID: 36869137
- PMCID: PMC9984395
- DOI: 10.1038/s42003-023-04582-y
Rickettsia felis DNA recovered from a child who lived in southern Africa 2000 years ago
Abstract
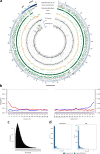
The Stone Age record of South Africa provides some of the earliest evidence for the biological and cultural origins of Homo sapiens. While there is extensive genomic evidence for the selection of polymorphisms in response to pathogen-pressure in sub-Saharan Africa, e.g., the sickle cell trait which provides protection against malaria, there is inadequate direct human genomic evidence for ancient human-pathogen infection in the region. Here, we analysed shotgun metagenome libraries derived from the sequencing of a Later Stone Age hunter-gatherer child who lived near Ballito Bay, South Africa, c. 2000 years ago. This resulted in the identification of ancient DNA sequence reads homologous to Rickettsia felis, the causative agent of typhus-like flea-borne rickettsioses, and the reconstruction of an ancient R. felis genome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests. The funding sponsors had no role in the design of the study, the collection, analyses, and interpretation of data, in the writing of the manuscript or in the decision to distribute the results.
Figures

References
-
- Lombard M, et al. Ancient human DNA: how sequencing the genome of a boy from Ballito Bay changed human history. S Afr. J. Sci. 2018;114:1–3. doi: 10.17159/sajs.2018/a0253. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

