Systematic analysis and expression of Gossypium 2ODD superfamily highlight the roles of GhLDOXs responding to alkali and other abiotic stress in cotton
- PMID: 36869319
- PMCID: PMC9985220
- DOI: 10.1186/s12870-023-04133-x
Systematic analysis and expression of Gossypium 2ODD superfamily highlight the roles of GhLDOXs responding to alkali and other abiotic stress in cotton
Abstract
Background: 2-oxoglutarate-dependent dioxygenase (2ODD) is the second largest family of oxidases involved in various oxygenation/hydroxylation reactions in plants. Many members in the family regulate gene transcription, nucleic acid modification/repair and secondary metabolic synthesis. The 2ODD family genes also function in the formation of abundant flavonoids during anthocyanin synthesis, thereby modulating plant development and response to diverse stresses.
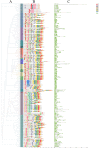
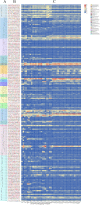
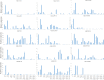
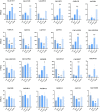
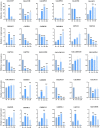
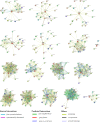
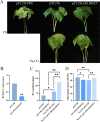
Results: Totally, 379, 336, 205, and 204 2ODD genes were identified in G. barbadense (Gb), G. hirsutum (Gh), G. arboreum (Ga), and G. raimondii (Gb), respectively. The 336 2ODDs in G. hirsutum were divided into 15 subfamilies according to their putative functions. The structural features and functions of the 2ODD members in the same subfamily were similar and evolutionarily conserved. Tandem duplications and segmental duplications served essential roles in the large-scale expansion of the cotton 2ODD family. Ka/Ks values for most of the gene pairs were less than 1, indicating that 2ODD genes undergo strong purifying selection during evolution. Gh2ODDs might act in cotton responses to different abiotic stresses. GhLDOX3 and GhLDOX7, two members of the GhLDOX subfamily from Gh2ODDs, were significantly down-regulated in transcription under alkaline stress. Moreover, the expression of GhLDOX3 in leaves was significantly higher than that in other tissues. These results will provide valuable information for further understanding the evolution mechanisms and functions of the cotton 2ODD genes in the future.
Conclusions: Genome-wide identification, structure, and evolution and expression analysis of 2ODD genes in Gossypium were carried out. The 2ODDs were highly conserved during evolutionary. Most Gh2ODDs were involved in the regulation of cotton responses to multiple abiotic stresses including salt, drought, hot, cold and alkali.
Keywords: Cotton; Gene family; Leucocyanidin dioxygenase; Phylogenetic analysis; Structural analysis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Genome-wide characterization and expression analysis of the aldehyde dehydrogenase (ALDH) gene superfamily under abiotic stresses in cotton.Gene. 2017 Sep 10;628:230-245. doi: 10.1016/j.gene.2017.07.034. Epub 2017 Jul 12. Gene. 2017. PMID: 28711668
-
Genome-wide identification and expression analysis of PUB genes in cotton.BMC Genomics. 2020 Mar 6;21(1):213. doi: 10.1186/s12864-020-6638-5. BMC Genomics. 2020. PMID: 32143567 Free PMC article.
-
Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species.BMC Genomics. 2017 Apr 12;18(1):292. doi: 10.1186/s12864-017-3682-x. BMC Genomics. 2017. PMID: 28403834 Free PMC article.
-
Genome wide identification, classification and functional characterization of heat shock transcription factors in cultivated and ancestral cottons (Gossypium spp.).Int J Biol Macromol. 2021 Jul 1;182:1507-1527. doi: 10.1016/j.ijbiomac.2021.05.016. Epub 2021 May 7. Int J Biol Macromol. 2021. PMID: 33965497
-
Genome-wide expression analysis of phospholipase A1 (PLA1) gene family suggests phospholipase A1-32 gene responding to abiotic stresses in cotton.Int J Biol Macromol. 2021 Dec 1;192:1058-1074. doi: 10.1016/j.ijbiomac.2021.10.038. Epub 2021 Oct 14. Int J Biol Macromol. 2021. PMID: 34656543 Review.
Cited by
-
Genome-Wide Identification and Cold Stress Response Mechanism of Barley Di19 Gene Family.Biology (Basel). 2025 May 6;14(5):508. doi: 10.3390/biology14050508. Biology (Basel). 2025. PMID: 40427698 Free PMC article.
-
The Antarctic moss 2-oxoglutarate/Fe(II)-dependent dioxygenases (Pn2-ODD2) enhanced the tolerance to drought and oxidative stress.BMC Plant Biol. 2025 Apr 28;25(1):549. doi: 10.1186/s12870-025-06578-8. BMC Plant Biol. 2025. PMID: 40295947 Free PMC article.
-
Metabolomics Combined with Transcriptomics Analysis Reveals the Regulation of Flavonoids in the Leaf Color Change of Acer truncatum Bunge.Int J Mol Sci. 2024 Dec 12;25(24):13325. doi: 10.3390/ijms252413325. Int J Mol Sci. 2024. PMID: 39769090 Free PMC article.
References
-
- Li J, Pu L, Han M, Zhu M, Zhang R, Xiang Y. Soil salinization research in China: advances and prospects. J Geog Sci. 2014;24(5):943–960. doi: 10.3389/fpls.2021.667458. - DOI
-
- Wang H, Takano T, Liu S. Screening and evaluation of saline–alkaline tolerant germplasm of rice (Oryza sativa L.) in soda saline–alkali soil. Agronomy. 2018;8(10):205. 10.3390/agronomy8100205.
-
- Wang X, Ren H, Wei Z, Wang Y, Ren W. Effects of neutral salt and alkali on ion distributions in the roots, shoots, and leaves of two alfalfa cultivars with differing degrees of salt tolerance. J Integr Agric. 2017;16(8):1800–07. doi: 10.1016/S2095-3119(16)61522-8. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

