Clinical and genomic features of non-small cell lung cancer occurring in families
- PMID: 36869602
- PMCID: PMC10067361
- DOI: 10.1111/1759-7714.14825
Clinical and genomic features of non-small cell lung cancer occurring in families
Abstract
Background: Exposure to environmental carcinogens, such as through smoking, is a major factor in the carcinogenesis of non-small cell lung cancer (NSCLC). However, genetic factors may also contribute.
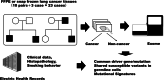
Methods: To identify candidate tumor suppressor genes for NSCLC, we included 23 patients (10 related pairs and 3 individuals) with NSCLC who had other NSCLC-affected first-degree relatives in a local hospital. Exome analyses for both germline and somatic (NSCLC specimens) DNA were performed for 17 cases. Germline exome data of these 17 cases revealed that most of the short variants were identical to the variants in 14KJPN (a Japanese reference genome panel of more than 14 000 individuals) and only a nonsynonymous variant in the DHODH gene, p.A347T, was shared between a pair of NSCLC patients in the same family. This variant is a known pathogenic variant of the gene for Miller syndrome.
Results: Somatic genetic alterations in the exome data of our samples showed frequent mutations in the EGFR and TP53 genes. Principal component analysis of the patterns of 96 types of single nucleotide variants (SNVs) suggested the existence of unique mechanisms inducing somatic SNVs in each family. Delineation of mutational signatures of the somatic SNVs with deconstructSigs for the pair of germline pathogenic DHODH variant-positive cases showed that the mutational signatures of these cases included SBS3 (homologous recombination repair defect), SBS6, 15 (DNA mismatch repair), and SBS7 (ultraviolet exposure), suggesting that disordered pyrimidine production causes increased errors in DNA repair systems in these cases.
Conclusion: Our results suggest the importance of the detailed collection of data on environmental exposure along with genetic information on NSCLC patients to identify the unique combinations that cause lung tumorigenesis in a particular family.
Keywords: DHODH; exome; mutational signature; non-small cell lung cancer; tumor suppressor gene.
© 2023 The Authors. Thoracic Cancer published by China Lung Oncology Group and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors do not have any conflict of interest to declare.
Figures





References
-
- Malhotra J, Malvezzi M, Negri E, La Vecchia C, Boffetta P. Risk factors for lung cancer worldwide. Eur Respir J. 2016;48:889–902. - PubMed
-
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. - PubMed
-
- Yokota J, Shiraishi K, Kohno T. Genetic basis for susceptibility to lung cancer: recent progress and future directions. Adv Cancer Res. 2010;109:51–72. - PubMed
-
- Kohno T, Kunitoh H, Shimada Y, Shiraishi K, Ishii Y, Goto K, et al. Individuals susceptible to lung adenocarcinoma defined by combined HLA‐DQA1 and TERT genotypes. Carcinogenesis. 2010;31:834–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

