Phosphorylation and ubiquitination of OsWRKY31 are integral to OsMKK10-2-mediated defense responses in rice
- PMID: 36869655
- PMCID: PMC10226564
- DOI: 10.1093/plcell/koad064
Phosphorylation and ubiquitination of OsWRKY31 are integral to OsMKK10-2-mediated defense responses in rice
Abstract
Mitogen-activated protein kinase (MPK) cascades play vital roles in plant innate immunity, growth, and development. Here, we report that the rice (Oryza sativa) transcription factor gene OsWRKY31 is a key component in a MPK signaling pathway involved in plant disease resistance in rice. We found that the activation of OsMKK10-2 enhances resistance against the rice blast pathogen Magnaporthe oryzae and suppresses growth through an increase in jasmonic acid and salicylic acid accumulation and a decrease of indole-3-acetic acid levels. Knockout of OsWRKY31 compromises the defense responses mediated by OsMKK10-2. OsMKK10-2 and OsWRKY31 physically interact, and OsWRKY31 is phosphorylated by OsMPK3, OsMPK4, and OsMPK6. Phosphomimetic OsWRKY31 has elevated DNA-binding activity and confers enhanced resistance to M. oryzae. In addition, OsWRKY31 stability is regulated by phosphorylation and ubiquitination via RING-finger E3 ubiquitin ligases interacting with WRKY 1 (OsREIW1). Taken together, our findings indicate that modification of OsWRKY31 by phosphorylation and ubiquitination functions in the OsMKK10-2-mediated defense signaling pathway.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. The authors declare no conflict of interests.
Figures

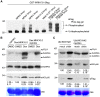





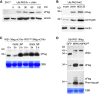

Comment in
-
WRKYng together: Coordination between kinase cascades and transcription factors contributes to immunity in rice.Plant Cell. 2023 May 29;35(6):1968-1969. doi: 10.1093/plcell/koad074. Plant Cell. 2023. PMID: 36911988 Free PMC article. No abstract available.
Similar articles
-
Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants.Cell Res. 2008 Apr;18(4):508-21. doi: 10.1038/cr.2007.104. Cell Res. 2008. PMID: 18071364
-
Molecular Dissection of Early Defense Signaling Underlying Volatile-Mediated Defense Regulation and Herbivore Resistance in Rice.Plant Cell. 2019 Mar;31(3):687-698. doi: 10.1105/tpc.18.00569. Epub 2019 Feb 13. Plant Cell. 2019. PMID: 30760558 Free PMC article.
-
A VQ-motif-containing protein fine-tunes rice immunity and growth by a hierarchical regulatory mechanism.Cell Rep. 2022 Aug 16;40(7):111235. doi: 10.1016/j.celrep.2022.111235. Cell Rep. 2022. PMID: 35977497
-
Current understanding of pattern-triggered immunity and hormone-mediated defense in rice (Oryza sativa) in response to Magnaporthe oryzae infection.Semin Cell Dev Biol. 2018 Nov;83:95-105. doi: 10.1016/j.semcdb.2017.10.020. Epub 2017 Nov 2. Semin Cell Dev Biol. 2018. PMID: 29061483 Review.
-
Ubiquitin signaling in immune responses.Cell Res. 2016 Apr;26(4):457-83. doi: 10.1038/cr.2016.40. Epub 2016 Mar 25. Cell Res. 2016. PMID: 27012466 Free PMC article. Review.
Cited by
-
E3 Ubiquitin Ligase OsRFI2 Regulates Salinity Tolerance by Targeting Ascorbate Peroxidase OsAPX8 for its Degradation in Rice.Rice (N Y). 2025 Mar 10;18(1):12. doi: 10.1186/s12284-025-00763-x. Rice (N Y). 2025. PMID: 40059282 Free PMC article.
-
Genome-Wide Identification of MKK Gene Family and Response to Hormone and Abiotic Stress in Rice.Plants (Basel). 2024 Oct 18;13(20):2922. doi: 10.3390/plants13202922. Plants (Basel). 2024. PMID: 39458871 Free PMC article.
-
WRKY Transcription Factors in Response to Metal Stress in Plants: A Review.Int J Mol Sci. 2024 Oct 11;25(20):10952. doi: 10.3390/ijms252010952. Int J Mol Sci. 2024. PMID: 39456735 Free PMC article. Review.
-
Post-Translational Modification of WRKY Transcription Factors.Plants (Basel). 2024 Jul 25;13(15):2040. doi: 10.3390/plants13152040. Plants (Basel). 2024. PMID: 39124158 Free PMC article. Review.
-
Multifaceted roles of WRKY transcription factors in abiotic stress and flavonoid biosynthesis.Front Plant Sci. 2023 Dec 15;14:1303667. doi: 10.3389/fpls.2023.1303667. eCollection 2023. Front Plant Sci. 2023. PMID: 38169626 Free PMC article. Review.
References
-
- Chen X, Li C, Wang H, Guo Z. WRKY transcription factors: evolution, binding, and action. Phytopathol Res. 2019:1(1): 13. 10.1186/s42483-019-0022-x - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

