Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER
- PMID: 36869664
- PMCID: PMC10085695
- DOI: 10.1093/nar/gkad108
Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER
Abstract
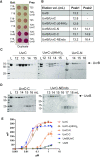
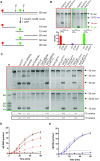
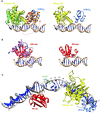
Bacterial nucleotide excision repair (NER), mediated by the UvrA, UvrB and UvrC proteins is a multistep, ATP-dependent process, that is responsible for the removal of a very wide range of chemically and structurally diverse DNA lesions. DNA damage removal is performed by UvrC, an enzyme possessing a dual endonuclease activity, capable of incising the DNA on either side of the damaged site to release a short single-stranded DNA fragment containing the lesion. Using biochemical and biophysical approaches, we have probed the oligomeric state, UvrB- and DNA-binding abilities and incision activities of wild-type and mutant constructs of UvrC from the radiation resistant bacterium, Deinococcus radiodurans. Moreover, by combining the power of new structure prediction algorithms and experimental crystallographic data, we have assembled the first model of a complete UvrC, revealing several unexpected structural motifs and in particular, a central inactive RNase H domain acting as a platform for the surrounding domains. In this configuration, UvrC is maintained in a 'closed' inactive state that needs to undergo a major rearrangement to adopt an 'open' active state capable of performing the dual incision reaction. Taken together, this study provides important insight into the mechanism of recruitment and activation of UvrC during NER.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Truglio J.J., Croteau D.L., Van Houten B., Kisker C.. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006; 106:233–252. - PubMed
-
- Jaciuk M., Swuec P., Gaur V., Kasprzak J.M., Renault L., Dobrychłop M., Nirwal S., Bujnicki J.M., Costa A., Nowotny M.. A combined structural and biochemical approach reveals translocation and stalling of UvrB on the DNA lesion as a mechanism of damage verification in bacterial nucleotide excision repair. DNA Repair (Amst.). 2020; 85:102746. - PMC - PubMed
-
- Pakotiprapha D., Jeruzalmi D.. Small-angle X-ray scattering reveals architecture and A(2)B(2) stoichiometry of the UvrA-UvrB DNA damage sensor. Proteins. 2013; 81:132–139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

