Fitness functions for RNA structure design
- PMID: 36869673
- PMCID: PMC10123107
- DOI: 10.1093/nar/gkad097
Fitness functions for RNA structure design
Abstract
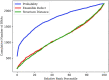
An RNA design algorithm takes a target RNA structure and finds a sequence that folds into that structure. This is fundamentally important for engineering therapeutics using RNA. Computational RNA design algorithms are guided by fitness functions, but not much research has been done on the merits of these functions. We survey current RNA design approaches with a particular focus on the fitness functions used. We experimentally compare the most widely used fitness functions in RNA design algorithms on both synthetic and natural sequences. It has been almost 20 years since the last comparison was published, and we find similar results with a major new result: maximizing probability outperforms minimizing ensemble defect. The probability is the likelihood of a structure at equilibrium and the ensemble defect is the weighted average number of incorrect positions in the ensemble. We find that maximizing probability leads to better results on synthetic RNA design puzzles and agrees more often than other fitness functions with natural sequences and structures, which were designed by evolution. Also, we observe that many recently published approaches minimize structure distance to the minimum free energy prediction, which we find to be a poor fitness function.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
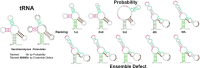

Similar articles
-
A statistical sampling algorithm for RNA secondary structure prediction.Nucleic Acids Res. 2003 Dec 15;31(24):7280-301. doi: 10.1093/nar/gkg938. Nucleic Acids Res. 2003. PMID: 14654704 Free PMC article.
-
Nucleic acid sequence design via efficient ensemble defect optimization.J Comput Chem. 2011 Feb;32(3):439-52. doi: 10.1002/jcc.21633. Epub 2010 Aug 17. J Comput Chem. 2011. PMID: 20717905
-
Efficient algorithms for probing the RNA mutation landscape.PLoS Comput Biol. 2008 Aug 8;4(8):e1000124. doi: 10.1371/journal.pcbi.1000124. PLoS Comput Biol. 2008. PMID: 18688270 Free PMC article.
-
Prediction and design of DNA and RNA structures.N Biotechnol. 2010 Jul 31;27(3):184-93. doi: 10.1016/j.nbt.2010.02.012. Epub 2010 Mar 1. N Biotechnol. 2010. PMID: 20193785 Review.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
Cited by
-
Context-dependent structure formation of hairpin motifs in bacteriophage MS2 genomic RNA.Biophys J. 2024 Oct 1;123(19):3397-3407. doi: 10.1016/j.bpj.2024.08.004. Epub 2024 Aug 8. Biophys J. 2024. PMID: 39118324
-
gRNAde: Geometric Deep Learning for 3D RNA inverse design.bioRxiv [Preprint]. 2025 Feb 26:2024.03.31.587283. doi: 10.1101/2024.03.31.587283. bioRxiv. 2025. Update in: Methods Mol Biol. 2025;2847:121-135. doi: 10.1007/978-1-0716-4079-1_8. PMID: 38826198 Free PMC article. Updated. Preprint.
-
Monte Carlo Inverse RNA Folding.Methods Mol Biol. 2025;2847:205-215. doi: 10.1007/978-1-0716-4079-1_14. Methods Mol Biol. 2025. PMID: 39312146
-
Analysis of natural structures and chemical mapping data reveals local stability compensation in RNA.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf565. doi: 10.1093/nar/gkaf565. Nucleic Acids Res. 2025. PMID: 40568944 Free PMC article.
-
Analysis of natural structures and chemical mapping data reveals local stability compensation in RNA.bioRxiv [Preprint]. 2025 Apr 11:2024.12.11.627843. doi: 10.1101/2024.12.11.627843. bioRxiv. 2025. Update in: Nucleic Acids Res. 2025 Jun 20;53(12):gkaf565. doi: 10.1093/nar/gkaf565. PMID: 39713387 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

