Downregulation of MALAT1 is a hallmark of tissue and peripheral proliferative T cells in COVID-19
- PMID: 36869729
- PMCID: PMC10243916
- DOI: 10.1093/cei/uxad034
Downregulation of MALAT1 is a hallmark of tissue and peripheral proliferative T cells in COVID-19
Abstract
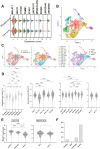
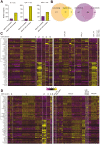
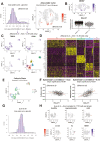
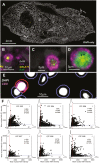
T cells play key protective but also pathogenic roles in COVID-19. We studied the expression of long non-coding RNAs (lncRNAs) in COVID-19 T-cell transcriptomes by integrating previously published single-cell RNA sequencing datasets. The long intergenic non-coding RNA MALAT1 was the most highly transcribed lncRNA in T cells, with Th1 cells demonstrating the lowest and CD8+ resident memory cells the highest MALAT1 expression, amongst CD4+ and CD8+ T-cells populations, respectively. We then identified gene signatures that covaried with MALAT1 in single T cells. A significantly higher number of transcripts correlated negatively with MALAT1 than those that correlated. Enriched functional annotations of the MALAT1- anti-correlating gene signature included processes associated with T-cell activation such as cell division, oxidative phosphorylation, and response to cytokine. The MALAT1 anti-correlating gene signature shared by both CD4+ and CD8+ T-cells marked dividing T cells in both the lung and blood of COVID-19 patients. Focussing on the tissue, we used an independent patient cohort of post-mortem COVID-19 lung samples and demonstrated that MALAT1 suppression was indeed a marker of MKI67+ proliferating CD8+ T cells. Our results reveal MALAT1 suppression and its associated gene signature are a hallmark of human proliferating T cells.
Keywords: COVID-19; MALAT1; T cell; lncRNA; proliferation.
© The Author(s) 2023. Published by Oxford University Press on behalf of the British Society for Immunology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

