Global Dynamics of Porcine Enteric Coronavirus PEDV Epidemiology, Evolution, and Transmission
- PMID: 36869744
- PMCID: PMC10027654
- DOI: 10.1093/molbev/msad052
Global Dynamics of Porcine Enteric Coronavirus PEDV Epidemiology, Evolution, and Transmission
Abstract
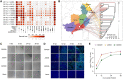
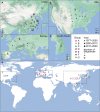
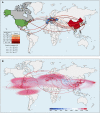
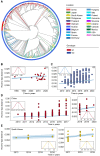
With a possible origin from bats, the alphacoronavirus Porcine epidemic diarrhea virus (PEDV) causes significant hazards and widespread epidemics in the swine population. However, the ecology, evolution, and spread of PEDV are still unclear. Here, from 149,869 fecal and intestinal tissue samples of pigs collected in an 11-year survey, we identified PEDV as the most dominant virus in diarrheal animals. Global whole genomic and evolutionary analyses of 672 PEDV strains revealed the fast-evolving PEDV genotype 2 (G2) strains as the main epidemic viruses worldwide, which seems to correlate with the use of G2-targeting vaccines. The evolving pattern of the G2 viruses presents geographic bias as they evolve tachytely in South Korea but undergo the highest recombination in China. Therefore, we clustered six PEDV haplotypes in China, whereas South Korea held five haplotypes, including a unique haplotype G. In addition, an assessment of the spatiotemporal spread route of PEDV indicates Germany and Japan as the primary hubs for PEDV dissemination in Europe and Asia, respectively. Overall, our findings provide novel insights into the epidemiology, evolution, and transmission of PEDV, and thus may lay a foundation for the prevention and control of PEDV and other coronaviruses.
Keywords: coronavirus; epidemiology; evolution; porcine epidemic diarrhea virus; transmission.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

