Biophysical forces mediated by respiration maintain lung alveolar epithelial cell fate
- PMID: 36870331
- PMCID: PMC10065960
- DOI: 10.1016/j.cell.2023.02.010
Biophysical forces mediated by respiration maintain lung alveolar epithelial cell fate
Abstract
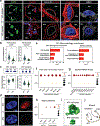
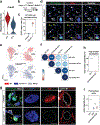
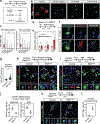
Lungs undergo mechanical strain during breathing, but how these biophysical forces affect cell fate and tissue homeostasis are unclear. We show that biophysical forces through normal respiratory motion actively maintain alveolar type 1 (AT1) cell identity and restrict these cells from reprogramming into AT2 cells in the adult lung. AT1 cell fate is maintained at homeostasis by Cdc42- and Ptk2-mediated actin remodeling and cytoskeletal strain, and inactivation of these pathways causes a rapid reprogramming into the AT2 cell fate. This plasticity induces chromatin reorganization and changes in nuclear lamina-chromatin interactions, which can discriminate AT1 and AT2 cell identity. Unloading the biophysical forces of breathing movements leads to AT1-AT2 cell reprogramming, revealing that normal respiration is essential to maintain alveolar epithelial cell fate. These data demonstrate the integral function of mechanotransduction in maintaining lung cell fate and identifies the AT1 cell as an important mechanosensor in the alveolar niche.
Keywords: alveolar epithelial cell; biophysical forces; lamina-associated domain; lung; mechanotransduction.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Comment in
-
Breathing-induced forces influence lung cell fate.Cell Stem Cell. 2023 May 4;30(5):507-508. doi: 10.1016/j.stem.2023.04.004. Cell Stem Cell. 2023. PMID: 37146577
References
-
- Liu F, Lagares D, Choi KM, Stopfer L, Marinkovic A, Vrbanac V, Probst CK, Hiemer SE, Sisson TH, Horowitz JC, et al. (2015). Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol 308, L344–357. 10.1152/ajplung.00300.2014. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

