Single molecule analyses reveal dynamics of Salmonella translocated effector proteins in host cell endomembranes
- PMID: 36870997
- PMCID: PMC9985595
- DOI: 10.1038/s41467-023-36758-9
Single molecule analyses reveal dynamics of Salmonella translocated effector proteins in host cell endomembranes
Abstract
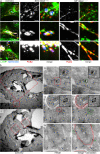
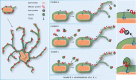
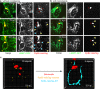
The facultative intracellular pathogen Salmonella enterica remodels the host endosomal system for survival and proliferation inside host cells. Salmonella resides within the Salmonella-containing vacuole (SCV) and by Salmonella-induced fusions of host endomembranes, the SCV is connected with extensive tubular structures termed Salmonella-induced filaments (SIF). The intracellular lifestyle of Salmonella critically depends on effector proteins translocated into host cells. A subset of effectors is associated with, or integral in SCV and SIF membranes. How effectors reach their subcellular destination, and how they interact with endomembranes remodeled by Salmonella remains to be determined. We deployed self-labeling enzyme tags to label translocated effectors in living host cells, and analyzed their single molecule dynamics. Translocated effectors diffuse in membranes of SIF with mobility comparable to membrane-integral host proteins in endomembranes. Dynamics differ between various effectors investigated and is dependent on membrane architecture of SIF. In the early infection, host endosomal vesicles are associated with Salmonella effectors. Effector-positive vesicles continuously fuse with SCV and SIF membranes, providing a route of effector delivery by translocation, interaction with endosomal vesicles, and ultimately fusion with the continuum of SCV/SIF membranes. This mechanism controls membrane deformation and vesicular fusion to generate the specific intracellular niche for bacterial survival and proliferation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

