NUDT22 promotes cancer growth through pyrimidine salvage
- PMID: 36871087
- PMCID: PMC10101856
- DOI: 10.1038/s41388-023-02643-4
NUDT22 promotes cancer growth through pyrimidine salvage
Abstract
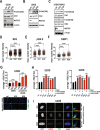
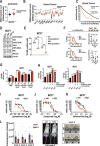
The NUDIX hydrolase NUDT22 converts UDP-glucose into glucose-1-phosphate and the pyrimidine nucleotide uridine monophosphate but a biological significance for this biochemical reaction has not yet been established. Glucose-1-phosphate is an important metabolite for energy and biomass production through glycolysis and nucleotides required for DNA replication are produced through energetically expensive de novo or energy-efficient salvage pathways. Here, we describe p53-regulated pyrimidine salvage through NUDT22-dependent hydrolysis of UDP-glucose to maintain cancer cell growth and to prevent replication stress. NUDT22 expression is consistently elevated in cancer tissues and high NUDT22 expression correlates with worse survival outcomes in patients indicating an increased dependency of cancer cells to NUDT22. Furthermore, we show that NUDT22 transcription is induced after inhibition of glycolysis, MYC-mediated oncogenic stress, and DNA damage directly through p53. NUDT22-deficient cancer cells suffer from growth retardation, S-phase delay, and slower DNA replication fork speed. Uridine supplementation rescues replication fork progression and alleviates replication stress and DNA damage. Conversely, NUDT22 deficiency sensitizes cells to de novo pyrimidine synthesis inhibition in vitro and reduces cancer growth in vivo. In conclusion, NUDT22 maintains pyrimidine supply in cancer cells and depletion of NUDT22 leads to genome instability. Targeting NUDT22 therefore has high potential for therapeutic applications in cancer therapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
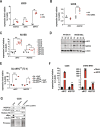

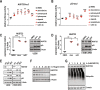
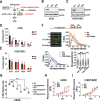
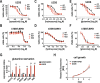
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

