Lipid Deposition Profiles Influence Foreign Body Responses
- PMID: 36871193
- PMCID: PMC10309593
- DOI: 10.1002/adma.202205709
Lipid Deposition Profiles Influence Foreign Body Responses
Abstract
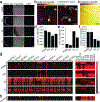
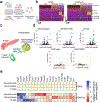
Fibrosis remains a significant cause of failure in implanted biomedical devices and early absorption of proteins on implant surfaces has been shown to be a key instigating factor. However, lipids can also regulate immune activity and their presence may also contribute to biomaterial-induced foreign body responses (FBR) and fibrosis. Here it is demonstrated that the surface presentation of lipids on implant affects FBR by influencing reactions of immune cells to materials as well as their resultant inflammatory/suppressive polarization. Time-of-flight secondary ion mass spectroscopy (ToF-SIMS) is employed to characterize lipid deposition on implants that are surface-modified chemically with immunomodulatory small molecules. Multiple immunosuppressive phospholipids (phosphatidylcholine, phosphatidylinositol, phosphatidylethanolamine, and sphingomyelin) are all found to deposit preferentially on implants with anti-FBR surface modifications in mice. Significantly, a set of 11 fatty acids is enriched on unmodified implanted devices that failed in both mice and humans, highlighting relevance across species. Phospholipid deposition is also found to upregulate the transcription of anti-inflammatory genes in murine macrophages, while fatty acid deposition stimulated the expression of pro-inflammatory genes. These results provide further insights into how to improve the design of biomaterials and medical devices to mitigate biomaterial material-induced FBR and fibrosis.
Keywords: biomaterials; foreign body responses; immune responses; lipids; medical devices; surface analyses; tof-sims.
© 2023 Wiley-VCH GmbH.
Conflict of interest statement
Conflicts of interest
C. Schreib, O. Veiseh, and S. Mukherjee declare interests via a patent filed by Rice University by the technologies developed in this manuscript. O. Veiseh declares conflict of interest through equity stake and is a paid consultant of Sigilon Therapeutics. The authors declare that they have no other competing interests.
Figures




References
-
- McKeen LW, in Handbook of Polymer Applications in Medicine and Medical Devices, Elsevier Inc., 2014, pp. 21–53.
-
- Rolfe B, Mooney J, Zhang B, Jahnke S, Le S-J, Chau Y-Q, Huang Q, Wang H, Campbell G, Campbell J, Regenerative Medicine and Tissue Engineering - Cells and Biomaterials 2011, DOI 10.5772/21790. - DOI
-
- Someya T, Bao Z, Malliaras GG, Nature 2016, 540, 379. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

