Intensity of sample processing methods impacts wastewater SARS-CoV-2 whole genome amplicon sequencing outcomes
- PMID: 36871720
- PMCID: PMC9984232
- DOI: 10.1016/j.scitotenv.2023.162572
Intensity of sample processing methods impacts wastewater SARS-CoV-2 whole genome amplicon sequencing outcomes
Abstract
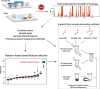
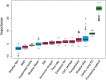
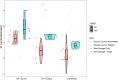
Wastewater SARS-CoV-2 surveillance has been deployed since the beginning of the COVID-19 pandemic to monitor the dynamics in virus burden in local communities. Genomic surveillance of SARS-CoV-2 in wastewater, particularly efforts aimed at whole genome sequencing for variant tracking and identification, are still challenging due to low target concentration, complex microbial and chemical background, and lack of robust nucleic acid recovery experimental procedures. The intrinsic sample limitations are inherent to wastewater and are thus unavoidable. Here, we use a statistical approach that couples correlation analyses to a random forest-based machine learning algorithm to evaluate potentially important factors associated with wastewater SARS-CoV-2 whole genome amplicon sequencing outcomes, with a specific focus on the breadth of genome coverage. We collected 182 composite and grab wastewater samples from the Chicago area between November 2020 to October 2021. Samples were processed using a mixture of processing methods reflecting different homogenization intensities (HA + Zymo beads, HA + glass beads, and Nanotrap), and were sequenced using one of the two library preparation kits (the Illumina COVIDseq kit and the QIAseq DIRECT kit). Technical factors evaluated using statistical and machine learning approaches include sample types, certain sample intrinsic features, and processing and sequencing methods. The results suggested that sample processing methods could be a predominant factor affecting sequencing outcomes, and library preparation kits was considered a minor factor. A synthetic SARS-CoV-2 RNA spike-in experiment was performed to validate the impact from processing methods and suggested that the intensity of the processing methods could lead to different RNA fragmentation patterns, which could also explain the observed inconsistency between qPCR quantification and sequencing outcomes. Overall, extra attention should be paid to wastewater sample processing (i.e., concentration and homogenization) for sufficient and good quality SARS-CoV-2 RNA for downstream sequencing.
Keywords: Amplicon sequencing; Illumina COVIDseq; QIAseq DIRECT; RNA fragmentation; Sample processing methods; Wastewater SARS-CoV-2.
Copyright © 2023. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Ahmed W., Bivins A., Korajkic A., Metcalfe S., Smith W.J.M., Simpson S.L. Comparative analysis of Adsorption-Extraction (AE) and Nanotrap® Magnetic Virus Particles (NMVP) workflows for the recovery of endogenous enveloped and non-enveloped viruses in wastewater. Sci. Total Environ. 2023;859 doi: 10.1016/j.scitotenv.2022.160072. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

