Actin-rich lamellipodia-like protrusions contribute to the integrity of epithelial cell-cell junctions
- PMID: 36871754
- PMCID: PMC10173786
- DOI: 10.1016/j.jbc.2023.104571
Actin-rich lamellipodia-like protrusions contribute to the integrity of epithelial cell-cell junctions
Abstract
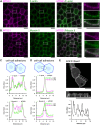
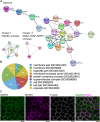
Metastasis-suppressor 1 (MTSS1) is a membrane-interacting scaffolding protein that regulates the integrity of epithelial cell-cell junctions and functions as a tumor suppressor in a wide range of carcinomas. MTSS1 binds phosphoinositide-rich membranes through its I-BAR domain and is capable of sensing and generating negative membrane curvature in vitro. However, the mechanisms by which MTSS1 localizes to intercellular junctions in epithelial cells and contributes to their integrity and maintenance have remained elusive. By carrying out EM and live-cell imaging on cultured Madin-Darby canine kidney cell monolayers, we provide evidence that adherens junctions of epithelial cells harbor lamellipodia-like, dynamic actin-driven membrane folds, which exhibit high negative membrane curvature at their distal edges. BioID proteomics and imaging experiments demonstrated that MTSS1 associates with an Arp2/3 complex activator, the WAVE-2 complex, in dynamic actin-rich protrusions at cell-cell junctions. Inhibition of Arp2/3 or WAVE-2 suppressed actin filament assembly at adherens junctions, decreased the dynamics of junctional membrane protrusions, and led to defects in epithelial integrity. Together, these results support a model in which membrane-associated MTSS1, together with the WAVE-2 and Arp2/3 complexes, promotes the formation of dynamic lamellipodia-like actin protrusions that contribute to the integrity of cell-cell junctions in epithelial monolayers.
Keywords: Arp2/3 complex; BAR domain; WAVE complex; actin; adherens junction; cell compartmentalization; epithelial cell; lipid-protein interaction; membrane curvature; protein self-assembly.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Senju Y., Lappalainen P. Regulation of actin dynamics by PI(4,5)P2 in cell migration and endocytosis. Curr. Opin. Cell Biol. 2019;56:7–13. - PubMed
-
- Gallop J.L. Filopodia and their links with membrane traffic and cell adhesion. Semin. Cell Dev. Biol. 2020;102:81–89. - PubMed
-
- Krause M., Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 2014;15:577–590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

