Honokiol alleviated neurodegeneration by reducing oxidative stress and improving mitochondrial function in mutant SOD1 cellular and mouse models of amyotrophic lateral sclerosis
- PMID: 36873166
- PMCID: PMC9979194
- DOI: 10.1016/j.apsb.2022.07.019
Honokiol alleviated neurodegeneration by reducing oxidative stress and improving mitochondrial function in mutant SOD1 cellular and mouse models of amyotrophic lateral sclerosis
Abstract
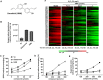
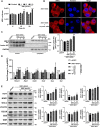
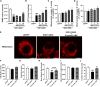
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease affecting both upper and lower motor neurons (MNs) with large unmet medical needs. Multiple pathological mechanisms are considered to contribute to the progression of ALS, including neuronal oxidative stress and mitochondrial dysfunction. Honokiol (HNK) has been reported to exert therapeutic effects in several neurologic disease models including ischemia stroke, Alzheimer's disease and Parkinson's disease. Here we found that honokiol also exhibited protective effects in ALS disease models both in vitro and in vivo. Honokiol improved the viability of NSC-34 motor neuron-like cells that expressed the mutant G93A SOD1 proteins (SOD1-G93A cells for short). Mechanistical studies revealed that honokiol alleviated cellular oxidative stress by enhancing glutathione (GSH) synthesis and activating the nuclear factor erythroid 2-related factor 2 (NRF2)-antioxidant response element (ARE) pathway. Also, honokiol improved both mitochondrial function and morphology via fine-tuning mitochondrial dynamics in SOD1-G93A cells. Importantly, honokiol extended the lifespan of the SOD1-G93A transgenic mice and improved the motor function. The improvement of antioxidant capacity and mitochondrial function was further confirmed in the spinal cord and gastrocnemius muscle in mice. Overall, honokiol showed promising preclinical potential as a multiple target drug for ALS treatment.
Keywords: Amyotrophic lateral sclerosis; Glutathione; Honokiol; Mitochondrial biogenesis; Mitochondrial dynamics; NRF2; Oxidative stress; SOD1-G93A.
© 2022 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures







References
-
- Pasinelli P., Brown R.H. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

