Discovery of lipid-mediated protein-protein interactions in living cells using metabolic labeling with photoactivatable clickable probes
- PMID: 36873846
- PMCID: PMC9977449
- DOI: 10.1039/d2sc06116c
Discovery of lipid-mediated protein-protein interactions in living cells using metabolic labeling with photoactivatable clickable probes
Abstract
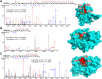
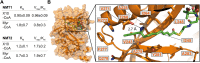
Protein-protein interactions (PPIs) are essential and pervasive regulatory elements in biology. Despite the development of a range of techniques to probe PPIs in living systems, there is a dearth of approaches to capture interactions driven by specific post-translational modifications (PTMs). Myristoylation is a lipid PTM added to more than 200 human proteins, where it may regulate membrane localization, stability or activity. Here we report the design and synthesis of a panel of novel photocrosslinkable and clickable myristic acid analog probes, and their characterization as efficient substrates for human N-myristoyltransferases NMT1 and NMT2, both biochemically and through X-ray crystallography. We demonstrate metabolic incorporation of probes to label NMT substrates in cell culture and in situ intracellular photoactivation to form a covalent crosslink between modified proteins and their interactors, capturing a snapshot of interactions in the presence of the lipid PTM. Proteomic analyses revealed both known and multiple novel interactors of a series of myristoylated proteins, including ferroptosis suppressor protein 1 (FSP1) and spliceosome-associated RNA helicase DDX46. The concept exemplified by these probes offers an efficient approach for exploring the PTM-specific interactome without the requirement for genetic modification, which may prove broadly applicable to other PTMs.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
EWT is a founder, shareholder and Director of Myricx Pharma Ltd.
Figures




References
-
- Walsh C. T., Posttranslational modification of proteins: Expanding nature's inventory, Greenwood Village, CO: Roberts and Company Publishers, 2005, p. 576
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

