Exploration and identification of anoikis-related genes in polycythemia vera
- PMID: 36873934
- PMCID: PMC9981965
- DOI: 10.3389/fgene.2023.1139351
Exploration and identification of anoikis-related genes in polycythemia vera
Abstract
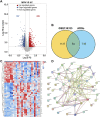
Background: Polycythemia Vera (PV) is a type of typical Myeloproliferative Neoplasms (MPNs) characterized with excessive erythropoiesis and thrombosis. Anoikis is a special programmed cell death mode induced by the adhesion disorder between cells and extracellular matrix (ECM) or adjacent cells facilitating cancer metastasis. However, few studies have focused on the role of anoikis in PV, especially on the development of PV. Methods: The microarray and RNA-seq results were screened from the Gene Expression Omnibus (GEO) database and the anoikis-related genes (ARGs) were downloaded from Genecards. The functional enrichment analysis of intersecting differentially expressed genes (DEGs) and protein-protein interaction (PPI) network analysis were performed to discover hub genes. The hub genes expression was tested in the training (GSE136335) and validation cohort (GSE145802), and RT-qPCR was performed to verify the gene expression in PV mice. Results: In the training GSE136335, a total of 1,195 DEGs was obtained from Myeloproliferative Neoplasm (MPN) patients compared with controls, among which 58 were anoikis-related DEGs. The significant enrichment of the apoptosis and cell adhesion pathways (i.e., cadherin binding) were shown in functional enrichment analysis. The PPI network was conducted to identify top five hub genes (CASP3, CYCS, HIF1A, IL1B, MCL1). The expression of CASP3 and IL1B were significantly upregulated both in validation cohort and PV mice and downregulated after treatment, suggesting that CASP3 and IL1B could be important indicators for disease surveillance. Conclusion: Our research revealed a relationship between anoikis and PV for the first time by combined analysis of gene level, protein interaction and functional enrichment, allowing novel insights into mechanisms of PV. Moreover, CASP3 and IL1B may become promising indicators of PV development and treatment.
Keywords: anoikis-related genes; differentially expressed genes; hub genes; myeloproliferative neoplasms; protein-protein interaction.
Copyright © 2023 Aini, Xie, Hu, Tang, Peng, Zhang and Deng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

