An integrated co-expression network analysis reveals novel genetic biomarkers for immune cell infiltration in chronic kidney disease
- PMID: 36875100
- PMCID: PMC9981626
- DOI: 10.3389/fimmu.2023.1129524
An integrated co-expression network analysis reveals novel genetic biomarkers for immune cell infiltration in chronic kidney disease
Abstract
Background: Chronic kidney disease (CKD) is characterized by persistent damage to kidney function or structure. Progression to end-stage leads to adverse effects on multiple systems. However, owing to its complex etiology and long-term cause, the molecular basis of CKD is not completely known.
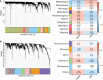
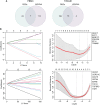
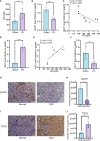
Methods: To dissect the potential important molecules during the progression, based on CKD databases from Gene Expression Omnibus, we used weighted gene co-expression network analysis (WGCNA) to identify the key genes in kidney tissues and peripheral blood mononuclear cells (PBMC). Correlation analysis of these genes with clinical relevance was evaluated based on Nephroseq. Combined with a validation cohort and receiver operating characteristic curve (ROC), we found the candidate biomarkers. The immune cell infiltration of these biomarkers was evaluated. The expression of these biomarkers was further detected in folic acid-induced nephropathy (FAN) murine model and immunohistochemical staining.
Results: In total, eight genes (CDCP1, CORO1C, DACH1, GSTA4, MAFB, TCF21, TGFBR3, and TGIF1) in kidney tissue and six genes (DDX17, KLF11, MAN1C1, POLR2K, ST14, and TRIM66) in PBMC were screened from co-expression network. Correlation analysis of these genes with serum creatinine levels and estimated glomerular filtration rate from Nephroseq showed a well clinical relevance. Validation cohort and ROC identified TCF21, DACH1 in kidney tissue and DDX17 in PBMC as biomarkers for the progression of CKD. Immune cell infiltration analysis revealed that DACH1 and TCF21 were correlated with eosinophil, activated CD8 T cell, activated CD4 T cell, while the DDX17 was correlated with neutrophil, type-2 T helper cell, type-1 T helper cell, mast cell, etc. FAN murine model and immunohistochemical staining confirmed that these three molecules can be used as genetic biomarkers to distinguish CKD patients from healthy people. Moreover, the increase of TCF21 in kidney tubules might play important role in the CKD progression.
Discussion: We identified three promising genetic biomarkers which could play important roles in the progression of CKD.
Keywords: CKD; DACH1; DDX17; PBMC; TCF21; WGCNA; biomarkers.
Copyright © 2023 Xia, Hou, Cai, Xu, Yang, Huang and Mou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Screening of immune-related secretory proteins linking chronic kidney disease with calcific aortic valve disease based on comprehensive bioinformatics analysis and machine learning.J Transl Med. 2023 Jun 1;21(1):359. doi: 10.1186/s12967-023-04171-x. J Transl Med. 2023. PMID: 37264340 Free PMC article.
-
The key candidate genes in tubulointerstitial injury of chronic kidney diseases patients as determined by bioinformatic analysis.Cell Biochem Funct. 2020 Aug;38(6):761-772. doi: 10.1002/cbf.3545. Epub 2020 Apr 27. Cell Biochem Funct. 2020. PMID: 32340064
-
Exploring diagnostic biomarkers of type 2 cardio-renal syndrome based on secreted proteins and bioinformatics analysis.Sci Rep. 2024 Oct 19;14(1):24612. doi: 10.1038/s41598-024-75580-1. Sci Rep. 2024. PMID: 39427047 Free PMC article.
-
Role of cytogenetic biomarkers in management of chronic kidney disease patients: A review.Int J Health Sci (Qassim). 2016 Oct;10(4):576-589. Int J Health Sci (Qassim). 2016. PMID: 27833523 Free PMC article. Review.
-
Review on Inflammation Markers in Chronic Kidney Disease.Biomedicines. 2021 Feb 11;9(2):182. doi: 10.3390/biomedicines9020182. Biomedicines. 2021. PMID: 33670423 Free PMC article. Review.
Cited by
-
Sesame Seeds: A Nutrient-Rich Superfood.Foods. 2024 Apr 10;13(8):1153. doi: 10.3390/foods13081153. Foods. 2024. PMID: 38672826 Free PMC article. Review.
-
Utilizing rat kidney gene co-expression networks to enhance safety assessment biomarker identification and human translation.iScience. 2025 Jun 21;28(7):112978. doi: 10.1016/j.isci.2025.112978. eCollection 2025 Jul 18. iScience. 2025. PMID: 40687826 Free PMC article.
-
The potential crosstalk genes and molecular mechanisms between systemic lupus erythematosus and periodontitis.Front Genet. 2025 Apr 16;16:1527713. doi: 10.3389/fgene.2025.1527713. eCollection 2025. Front Genet. 2025. PMID: 40309038 Free PMC article.
-
[Upregulating KLF11 ameliorates intestinal inflammation in mice with 2, 4, 6-trinitrobenesulfonic acid-induced colitis by inhibiting the JAK2/STAT3 signaling pathway].Nan Fang Yi Ke Da Xue Xue Bao. 2024 Apr 20;44(4):765-772. doi: 10.12122/j.issn.1673-4254.2024.04.19. Nan Fang Yi Ke Da Xue Xue Bao. 2024. PMID: 38708511 Free PMC article. Chinese.
-
Epigenome-wide association study of Chinese monozygotic twins identifies DNA methylation loci associated with estimated glomerular filtration rate.J Transl Med. 2025 Jan 22;23(1):101. doi: 10.1186/s12967-025-06067-4. J Transl Med. 2025. PMID: 39844292 Free PMC article.
References
-
- Inrig JK, Califf RM, Tasneem A, Vegunta RK, Molina C, Stanifer JW, et al. . The landscape of clinical trials in nephrology: a systematic review of clinicaltrials.gov. Am J Kidney Dis (2014) 63(5):771–80. doi: 10.1053/j.ajkd.2013.10.043 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

