Virulence network of interacting domains of influenza a and mouse proteins
- PMID: 36875146
- PMCID: PMC9982101
- DOI: 10.3389/fbinf.2023.1123993
Virulence network of interacting domains of influenza a and mouse proteins
Abstract
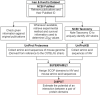
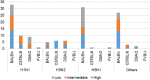
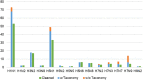
There exist several databases that provide virus-host protein interactions. While most provide curated records of interacting virus-host protein pairs, information on the strain-specific virulence factors or protein domains involved, is lacking. Some databases offer incomplete coverage of influenza strains because of the need to sift through vast amounts of literature (including those of major viruses including HIV and Dengue, besides others). None have offered complete, strain specific protein-protein interaction records for the influenza A group of viruses. In this paper, we present a comprehensive network of predicted domain-domain interaction(s) (DDI) between influenza A virus (IAV) and mouse host proteins, that will allow the systematic study of disease factors by taking the virulence information (lethal dose) into account. From a previously published dataset of lethal dose studies of IAV infection in mice, we constructed an interacting domain network of mouse and viral protein domains as nodes with weighted edges. The edges were scored with the Domain Interaction Statistical Potential (DISPOT) to indicate putative DDI. The virulence network can be easily navigated via a web browser, with the associated virulence information (LD50 values) prominently displayed. The network will aid influenza A disease modeling by providing strain-specific virulence levels with interacting protein domains. It can possibly contribute to computational methods for uncovering influenza infection mechanisms mediated through protein domain interactions between viral and host proteins. It is available at https://iav-ppi.onrender.com/home.
Keywords: domain-domain interaction; influenza a; lethal dose 50; mouse model; protein; virulence.
Copyright © 2023 Ng, Rashid and Kwoh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
A systems biology-driven approach to construct a comprehensive protein interaction network of influenza A virus with its host.BMC Infect Dis. 2020 Jul 6;20(1):480. doi: 10.1186/s12879-020-05214-0. BMC Infect Dis. 2020. PMID: 32631335 Free PMC article.
-
Rule-based meta-analysis reveals the major role of PB2 in influencing influenza A virus virulence in mice.BMC Genomics. 2019 Dec 24;20(Suppl 9):973. doi: 10.1186/s12864-019-6295-8. BMC Genomics. 2019. PMID: 31874643 Free PMC article.
-
Virulence determinants of avian H5N1 influenza A virus in mammalian and avian hosts: role of the C-terminal ESEV motif in the viral NS1 protein.J Virol. 2010 Oct;84(20):10708-18. doi: 10.1128/JVI.00610-10. Epub 2010 Aug 4. J Virol. 2010. PMID: 20686040 Free PMC article.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
The role of fusion activity of influenza A viruses in their biological properties.Acta Virol. 2016 Jun;60(2):121-35. doi: 10.4149/av_2016_02_121. Acta Virol. 2016. PMID: 27265461 Review.
Cited by
-
Identification of RC3H1 as antiviral host factor binding to the non-structural protein 1 of Influenza A virus via a 3-stage computational pipeline and cell-based analysis.Virol J. 2025 Apr 26;22(1):119. doi: 10.1186/s12985-025-02746-2. Virol J. 2025. PMID: 40287742 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

