Molecular acclimation of Halobacterium salinarum to halite brine inclusions
- PMID: 36875534
- PMCID: PMC9976938
- DOI: 10.3389/fmicb.2022.1075274
Molecular acclimation of Halobacterium salinarum to halite brine inclusions
Abstract
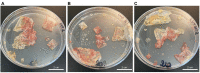
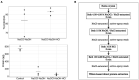
Halophilic microorganisms have long been known to survive within the brine inclusions of salt crystals, as evidenced by the change in color for salt crystals containing pigmented halophiles. However, the molecular mechanisms allowing this survival has remained an open question for decades. While protocols for the surface sterilization of halite (NaCl) have enabled isolation of cells and DNA from within halite brine inclusions, "-omics" based approaches have faced two main technical challenges: (1) removal of all contaminating organic biomolecules (including proteins) from halite surfaces, and (2) performing selective biomolecule extractions directly from cells contained within halite brine inclusions with sufficient speed to avoid modifications in gene expression during extraction. In this study, we tested different methods to resolve these two technical challenges. Following this method development, we then applied the optimized methods to perform the first examination of the early acclimation of a model haloarchaeon (Halobacterium salinarum NRC-1) to halite brine inclusions. Examinations of the proteome of Halobacterium cells two months post-evaporation revealed a high degree of similarity with stationary phase liquid cultures, but with a sharp down-regulation of ribosomal proteins. While proteins for central metabolism were part of the shared proteome between liquid cultures and halite brine inclusions, proteins involved in cell mobility (archaellum, gas vesicles) were either absent or less abundant in halite samples. Proteins unique to cells within brine inclusions included transporters, suggesting modified interactions between cells and the surrounding brine inclusion microenvironment. The methods and hypotheses presented here enable future studies of the survival of halophiles in both culture model and natural halite systems.
Keywords: Halobacterium; LC-MS; halite (NaCl); halophile; proteomics.
Copyright © 2023 Favreau, Tribondeau, Marugan, Guyot, Alpha-Bazin, Marie, Puppo, Dufour, Huguet, Zirah and Kish.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials

