RNA epitranscriptomics dysregulation: A major determinant for significantly increased risk of ASD pathogenesis
- PMID: 36875672
- PMCID: PMC9978375
- DOI: 10.3389/fnins.2023.1101422
RNA epitranscriptomics dysregulation: A major determinant for significantly increased risk of ASD pathogenesis
Abstract
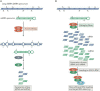
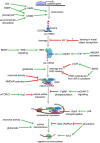
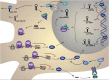
Autism spectrum disorders (ASDs) are perhaps the most severe, intractable and challenging child psychiatric disorders. They are complex, pervasive and highly heterogeneous and depend on multifactorial neurodevelopmental conditions. Although the pathogenesis of autism remains unclear, it revolves around altered neurodevelopmental patterns and their implications for brain function, although these cannot be specifically linked to symptoms. While these affect neuronal migration and connectivity, little is known about the processes that lead to the disruption of specific laminar excitatory and inhibitory cortical circuits, a key feature of ASD. It is evident that ASD has multiple underlying causes and this multigenic condition has been considered to also dependent on epigenetic effects, although the exact nature of the factors that could be involved remains unclear. However, besides the possibility for differential epigenetic markings directly affecting the relative expression levels of individual genes or groups of genes, there are at least three mRNA epitranscriptomic mechanisms, which function cooperatively and could, in association with both genotypes and environmental conditions, alter spatiotemporal proteins expression patterns during brain development, at both quantitative and qualitative levels, in a tissue-specific, and context-dependent manner. As we have already postulated, sudden changes in environmental conditions, such as those conferred by maternal inflammation/immune activation, influence RNA epitranscriptomic mechanisms, with the combination of these processes altering fetal brain development. Herein, we explore the postulate whereby, in ASD pathogenesis, RNA epitranscriptomics might take precedence over epigenetic modifications. RNA epitranscriptomics affects real-time differential expression of receptor and channel proteins isoforms, playing a prominent role in central nervous system (CNS) development and functions, but also RNAi which, in turn, impact the spatiotemporal expression of receptors, channels and regulatory proteins irrespective of isoforms. Slight dysregulations in few early components of brain development, could, depending upon their extent, snowball into a huge variety of pathological cerebral alterations a few years after birth. This may very well explain the enormous genetic, neuropathological and symptomatic heterogeneities that are systematically associated with ASD and psychiatric disorders at large.
Keywords: Autism spectrum disorder (ASD); RNA epitranscriptomics; adenosine-to-inosine RNA editing; mRNA alternative splicing; mRNA poly(A)-tail modulation; maternal inflammation.
Copyright © 2023 Beopoulos, Géa, Fasano and Iris.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








Similar articles
-
Autism spectrum disorders pathogenesis: Toward a comprehensive model based on neuroanatomic and neurodevelopment considerations.Front Neurosci. 2022 Nov 3;16:988735. doi: 10.3389/fnins.2022.988735. eCollection 2022. Front Neurosci. 2022. PMID: 36408388 Free PMC article.
-
Neuropsychopathology of Autism Spectrum Disorder: Complex Interplay of Genetic, Epigenetic, and Environmental Factors.Adv Neurobiol. 2020;24:97-141. doi: 10.1007/978-3-030-30402-7_4. Adv Neurobiol. 2020. PMID: 32006358 Review.
-
Dynamic changes in spatiotemporal transcriptome reveal maternal immune dysregulation of autism spectrum disorder.Comput Biol Med. 2022 Dec;151(Pt B):106334. doi: 10.1016/j.compbiomed.2022.106334. Epub 2022 Nov 17. Comput Biol Med. 2022. PMID: 36442276
-
Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder.Mol Psychiatry. 2018 Apr;23(4):1001-1013. doi: 10.1038/mp.2017.15. Epub 2017 Mar 21. Mol Psychiatry. 2018. PMID: 28322282 Free PMC article.
-
Variation in Gene Expression in Autism Spectrum Disorders: An Extensive Review of Transcriptomic Studies.Front Neurosci. 2017 Jan 5;10:601. doi: 10.3389/fnins.2016.00601. eCollection 2016. Front Neurosci. 2017. PMID: 28105001 Free PMC article. Review.
Cited by
-
Targeted drug screening for autism based on Cav1.2 calcium ion channel.PLoS One. 2025 May 29;20(5):e0324018. doi: 10.1371/journal.pone.0324018. eCollection 2025. PLoS One. 2025. PMID: 40440280 Free PMC article.
-
Improving Functional Abilities in Children and Adolescents with Autism Spectrum Disorder Using Non-Invasive REAC Neuro Psycho Physical Optimization Treatments: A PEDI-CAT Study.J Pers Med. 2023 May 5;13(5):792. doi: 10.3390/jpm13050792. J Pers Med. 2023. PMID: 37240963 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

