Generation and characterization of NGLY1 patient-derived midbrain organoids
- PMID: 36875753
- PMCID: PMC9978932
- DOI: 10.3389/fcell.2023.1039182
Generation and characterization of NGLY1 patient-derived midbrain organoids
Abstract
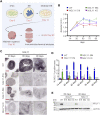
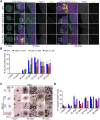
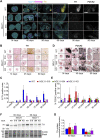
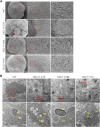
NGLY1 deficiency is an ultra-rare, autosomal recessive genetic disease caused by mutations in the NGLY1 gene encoding N-glycanase one that removes N-linked glycan. Patients with pathogenic mutations in NGLY1 have complex clinical symptoms including global developmental delay, motor disorder and liver dysfunction. To better understand the disease pathogenesis and the neurological symptoms of the NGLY1 deficiency we generated and characterized midbrain organoids using patient-derived iPSCs from two patients with distinct disease-causing mutations-one homozygous for p. Q208X, the other compound heterozygous for p. L318P and p. R390P and CRISPR generated NGLY1 knockout iPSCs. We demonstrate that NGLY1 deficient midbrain organoids show altered neuronal development compared to one wild type (WT) organoid. Both neuronal (TUJ1) and astrocytic glial fibrillary acid protein markers were reduced in NGLY1 patient-derived midbrain organoids along with neurotransmitter GABA. Interestingly, staining for dopaminergic neuronal marker, tyrosine hydroxylase, revealed a significant reduction in patient iPSC derived organoids. These results provide a relevant NGLY1 disease model to investigate disease mechanisms and evaluate therapeutics for treatments of NGLY1 deficiency.
Keywords: GABA; GFAP; NGLY1; dopaminergic neurons; midbrain organoids.
Copyright © 2023 Abbott, Tambe, Pavlinov, Farkhondeh, Nguyen, Xu, Pradhan, York, Might, Baumgärtel, Rodems and Zheng.
Conflict of interest statement
Author TY was employed by NeuroScience Associates Inc. Authors KB and SR were employed by Travere Therapeutics. Author HNN was employed by 3Dnamics Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

