Insights into the ecological generalist lifestyle of Clonostachys fungi through analysis of their predicted secretomes
- PMID: 36876087
- PMCID: PMC9978495
- DOI: 10.3389/fmicb.2023.1112673
Insights into the ecological generalist lifestyle of Clonostachys fungi through analysis of their predicted secretomes
Abstract
Introduction: The fungal secretome comprise diverse proteins that are involved in various aspects of fungal lifestyles, including adaptation to ecological niches and environmental interactions. The aim of this study was to investigate the composition and activity of fungal secretomes in mycoparasitic and beneficial fungal-plant interactions.
Methods: We used six Clonostachys spp. that exhibit saprotrophic, mycotrophic and plant endophytic lifestyles. Genome-wide analyses was performed to investigate the composition, diversity, evolution and gene expression of Clonostachys secretomes in relation to their potential role in mycoparasitic and endophytic lifestyles.
Results and discussion: Our analyses showed that the predicted secretomes of the analyzed species comprised between 7 and 8% of the respective proteomes. Mining of transcriptome data collected during previous studies showed that 18% of the genes encoding predicted secreted proteins were upregulated during the interactions with the mycohosts Fusarium graminearum and Helminthosporium solani. Functional annotation of the predicted secretomes revealed that the most represented protease family was subclass S8A (11-14% of the total), which include members that are shown to be involved in the response to nematodes and mycohosts. Conversely, the most numerous lipases and carbohydrate-active enzyme (CAZyme) groups appeared to be potentially involved in eliciting defense responses in the plants. For example, analysis of gene family evolution identified nine CAZyme orthogroups evolving for gene gains (p ≤ 0.05), predicted to be involved in hemicellulose degradation, potentially producing plant defense-inducing oligomers. Moreover, 8-10% of the secretomes was composed of cysteine-enriched proteins, including hydrophobins, important for root colonization. Effectors were more numerous, comprising 35-37% of the secretomes, where certain members belonged to seven orthogroups evolving for gene gains and were induced during the C. rosea response to F. graminearum or H. solani. Furthermore, the considered Clonostachys spp. possessed high numbers of proteins containing Common in Fungal Extracellular Membranes (CFEM) modules, known for their role in fungal virulence. Overall, this study improves our understanding of Clonostachys spp. adaptation to diverse ecological niches and establishes a basis for future investigation aiming at sustainable biocontrol of plant diseases.
Keywords: CFEM proteins; Clonostachys; antagonism; biocontrol; effector; mycoparasitism; secretome; small secreted cysteine-rich proteins.
Copyright © 2023 Piombo, Guaschino, Jensen, Karlsson and Dubey.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

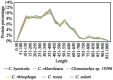



References
-
- Ahman J., Johansson T., Olsson M., Punt P. J., van den Hondel C. A., Tunlid A. (2002). Improving the pathogenicity of a nematode-trapping fungus by genetic engineering of a subtilisin with nematotoxic activity. Appl. Environ. Microbiol. 68, 3408–3415. doi: 10.1128/AEM.68.7.3408-3415.2002, PMID: - DOI - PMC - PubMed
-
- Alvindia D. G., Natsuaki K. T. (2008). Evaluation of fungal epiphytes isolated from banana fruit surfaces for biocontrol of banana crown rot disease. Crop Prot. 27, 1200–1207. doi: 10.1016/j.cropro.2008.02.007 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

