Differential prognosis of single and multiple TP53 abnormalities in high-count MBL and untreated CLL
- PMID: 36877634
- PMCID: PMC10338209
- DOI: 10.1182/bloodadvances.2022009040
Differential prognosis of single and multiple TP53 abnormalities in high-count MBL and untreated CLL
Abstract
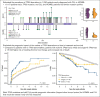
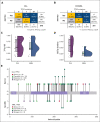
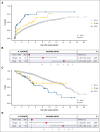
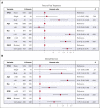
TP53 aberrations, including mutations and deletion of 17p13, are important adverse prognostic markers in chronic lymphocytic leukemia (CLL) but are less studied in high count monoclonal B-cell lymphocytosis (HCMBL), an asymptomatic pre-malignant stage of CLL. Here we estimated the prevalence and impact of TP53 aberrations in 1,230 newly diagnosed treatment-naïve individuals (849 CLL, 381 HCMBL). We defined TP53 state as: wild-type (no TP53 mutations and normal 17p), single-hit (del(17p) or one TP53 mutation), or multi-hit (TP53 mutation and del(17p), TP53 mutation and loss of heterozygosity, or multiple TP53 mutations). Cox regression was used to estimate hazard ratios (HR) and 95% confidence intervals (CI) for time to first treatment and overall survival by TP53 state. We found 64 (7.5%) CLL patients and 17 (4.5%) HCMBL individuals had TP53 mutations with variant allele fraction >10%. Del(17p) was present in 58 (6.8%) of CLL and 11 (2.9%) of HCMBL cases. Most individuals had wild-type (N=1,128, 91.7%) TP53 state, followed by multi-hit (N=55, 4.5%) and then single-hit (N=47, 3.8%) TP53 state. The risk of shorter time to therapy and death increased with the number of TP53 abnormalities. Compared to wild-type patients, multi-hit patients had 3-fold and single-hit patients had 1.5-fold increased risk of requiring therapy. Multi-hit patients also had 2.9-fold increased risk of death compared to wild-type. These results remained stable after accounting for other known poor prognostic factors. Both TP53 mutations and del(17p) may provide important prognostic information for HCMBL and CLL that would be missed if only one were measured.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: N.E.K. serves on the advisory board for AbbVie, AstraZeneca, Beigene, Behring, Boehringer Ingelheim Pharmaceuticals Inc, Cytomx Therapy, Dava Oncology, Janssen, Juno Therapeutics, Oncotracker, Pharmacyclics, and Targeted Oncology; serves on the data safety monitoring committee for Agios Pharmaceuticals, AstraZeneca, Bristol Myers Squibb, Celgene, CytomX Therapeutics, Dren Bio, Janssen, MorphoSys, and Rigel; and has received research funding from AbbVie, Acerta Pharma, Bristol Myers Squibb, Celgene, Genentech, MEI Pharma, Pharmacyclics, Sunesis, TG Therapeutics, and Tolero Pharmaceuticals. The remaining authors declare no competing financial interests.
Figures
References
-
- Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. - PubMed
-
- Puente XS, Bea S, Valdes-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519–524. - PubMed
-
- Dicker F, Herholz H, Schnittger S, et al. The detection of TP53 mutations in chronic lymphocytic leukemia independently predicts rapid disease progression and is highly correlated with a complex aberrant karyotype. Leukemia. 2009;23(1):117–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

