Assembly and regulation of the mammalian mRNA processing body
- PMID: 36877681
- PMCID: PMC9987799
- DOI: 10.1371/journal.pone.0282496
Assembly and regulation of the mammalian mRNA processing body
Abstract
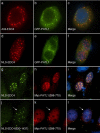
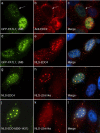
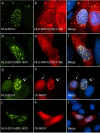
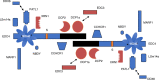
Messenger RNA processing bodies (P-bodies) are cytoplasmic membrane-free organelles that contain proteins involved in mRNA silencing, storage and decay. The mechanism by which P-body components interact and the factors that regulate the stability of these structures are incompletely understood. In this study, we used a fluorescence-based, two-hybrid assay to investigate interactions between P-body components that occur inside the cell. LSm14a, PATL1, XRN1, and NBDY were found to interact with the N-terminal, WD40-domain-containing portion of EDC4. The N-terminus of full-length PATL1 was required to mediate the interaction between EDC4 and DDX6. The C-terminal, alpha helix-domain- containing portion of EDC4 was sufficient to mediate interaction with DCP1a and CCHCR1. In the absence of endogenous P-bodies, caused by depletion of LSm14a or DDX6, expression of the portion of EDC4 that lacked the N-terminus retained the ability to form cytoplasmic dots that were indistinguishable from P-bodies at the level of UV light microscopy. Despite the absence of endogenous P-bodies, this portion of EDC4 was able to recruit DCP1a, CCHCR1 and EDC3 to cytoplasmic dots. The results of this study permit the development of a new model of P-body formation and suggest that the N-terminus of EDC4 regulates the stability of these structures.
Copyright: © 2023 Bloch et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

