CDK7 controls E2F- and MYC-driven proliferative and metabolic vulnerabilities in multiple myeloma
- PMID: 36877894
- PMCID: PMC10315622
- DOI: 10.1182/blood.2022018885
CDK7 controls E2F- and MYC-driven proliferative and metabolic vulnerabilities in multiple myeloma
Abstract
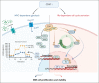
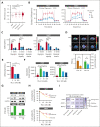
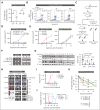
Therapeutic targeting of CDK7 has proven beneficial in preclinical studies, yet the off-target effects of currently available CDK7 inhibitors make it difficult to pinpoint the exact mechanisms behind MM cell death mediated by CDK7 inhibition. Here, we show that CDK7 expression positively correlates with E2F and MYC transcriptional programs in cells from patients with multiple myeloma (MM); its selective targeting counteracts E2F activity via perturbation of the cyclin-dependent kinases/Rb axis and impairs MYC-regulated metabolic gene signatures translating into defects in glycolysis and reduced levels of lactate production in MM cells. CDK7 inhibition using the covalent small-molecule inhibitor YKL-5-124 elicits a strong therapeutic response with minimal effects on normal cells, and causes in vivo tumor regression, increasing survival in several mouse models of MM including a genetically engineered mouse model of MYC-dependent MM. Through its role as a critical cofactor and regulator of MYC and E2F activity, CDK7 is therefore a master regulator of oncogenic cellular programs supporting MM growth and survival, and a valuable therapeutic target providing rationale for development of YKL-5-124 for clinical use.
Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution.
Conflict of interest statement
Conflict-of-interest disclosure: N.K. is now an employee of Kymera Therapeutics, Inc. N.S.G. is a founder, science advisory board member, and equity holder in Syros, C4, Allorion, Jengu, B2S, Inception, EoCys, Larkspur (board member), and Soltego (board member). The Gray laboratory receives, or has received, research funding from Novartis, Takeda, Astellas, Taiho, Janssen, Kinogen, Arbella, Deerfield, and Sanofi. N.S.G. and N.K. are named inventors on patent claiming YKL-5-124. K.C.A. has received consulting fees from Bristol-Myers Squibb (BMS), Celgene, Gilead, Janssen, Precision Biosciences, Sanofi-Aventis, Takeda, and Tolero; and serves on the board of directors of, and has stock options in, Oncopep. N.C.M. is a consultant for BMS, Janssen, Oncopep, Amgen, Karyopharm, Legened, AbbVie, Takeda, and GSK; and serves on the board of directors of, and stock options in, Oncopep. C.Y.L. is an executive and shareholder of Kronos Bio; has licensed intellectual property to Syros Pharmaceuticals; and is a shareholder and adviser of Ovibio, which is involved in the commercialization of YKL-5-124. R.A.Y. is a founder and equity holder in Syros Pharmaceuticals. B.N. is an inventor on patent applications related to the dTAG system (WO/2017/024318, WO/2017/024319, WO/2018/148440, WO/2018/148443, and WO/2020/146250). M.C. has received royalties for the Vk∗MYCDLox technology. C.M. serves on the scientific advisory board of Adicet Bio; and discloses consultant/honoraria from Genentech, Fate Therapeutics, Ionis Pharmaceuticals, FIMECS, Secura Bio, and Oncopeptides; and discloses research funding from Janssen/Johnson & Johnson, EMD Serono, Arch Oncology, Karyopharm, Sanofi, Nurix, BMS, H3 Biomedicine/Eisai, Springworks, Abcuro, and Novartis. The remaining authors declare no competing financial interests.
Figures



Comment in
-
Starving multiple myeloma cells via CDK7 inhibition.Blood. 2023 Jun 8;141(23):2787-2788. doi: 10.1182/blood.2023020254. Blood. 2023. PMID: 37289477 No abstract available.
References
-
- Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene. 2005;24(17):2909–2915. - PubMed
-
- Ramanathan Y, Rajpara SM, Reza SM, et al. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J Biol Chem. 2001;276(14):10913–10920. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

