Increased retinoic acid signaling decreases lung metastasis in salivary adenoid cystic carcinoma by inhibiting the noncanonical Notch1 pathway
- PMID: 36879115
- PMCID: PMC10073150
- DOI: 10.1038/s12276-023-00957-7
Increased retinoic acid signaling decreases lung metastasis in salivary adenoid cystic carcinoma by inhibiting the noncanonical Notch1 pathway
Abstract
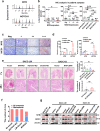
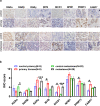
MYB-NFIB fusion and NOTCH1 mutation are common hallmark genetic events in salivary gland adenoid cystic carcinoma (SACC). However, abnormal expression of MYB and NOTCH1 is also observed in patients without MYB-NFIB fusion and NOTCH1 mutation. Here, we explore in-depth the molecular mechanisms of lung metastasis through single-cell RNA sequencing (scRNA-seq) and exome target capture sequencing in two SACC patients without MYB-NFIB fusion and NOTCH1 mutation. Twenty-five types of cells in primary and metastatic tissues were identified via Seurat clustering and categorized into four main stages ranging from near-normal to cancer-based on the abundance of each cell cluster in normal tissue. In this context, we identified the Notch signaling pathway enrichment in almost all cancer cells; RNA velocity, trajectory, and sub-clustering analyses were performed to deeply investigate cancer progenitor-like cell clusters in primary tumor-associated lung metastases, and signature genes of progenitor-like cells were enriched in the "MYC_TARGETS_V2" gene set. In vitro, we detected the NICD1-MYB-MYC complex by co-immunoprecipitation (Co-IP) and incidentally identified retinoic acid (RA) as an endogenous antagonist of genes in the "MYC_TARGETS_V2" gene set. Following this, we confirmed that all-trans retinoic acid (ATRA) suppresses the lung metastasis of SACC by correcting erroneous cell differentiation mainly caused by aberrant NOTCH1 or MYB expression. Bioinformatic, RNA-seq, and immunohistochemical (IHC) analyses of primary tissues and metastatic lung tissues from patients with SACC suggested that RA system insufficiency partially promotes lung metastasis. These findings imply the value of the RA system in diagnosis and treatment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

