Multiomic analyses implicate a neurodevelopmental program in the pathogenesis of cerebral arachnoid cysts
- PMID: 36879130
- PMCID: PMC12178143
- DOI: 10.1038/s41591-023-02238-2
Multiomic analyses implicate a neurodevelopmental program in the pathogenesis of cerebral arachnoid cysts
Abstract
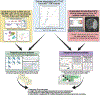
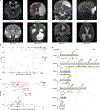
Cerebral arachnoid cysts (ACs) are one of the most common and poorly understood types of developmental brain lesion. To begin to elucidate AC pathogenesis, we performed an integrated analysis of 617 patient-parent (trio) exomes, 152,898 human brain and mouse meningeal single-cell RNA sequencing transcriptomes and natural language processing data of patient medical records. We found that damaging de novo variants (DNVs) were highly enriched in patients with ACs compared with healthy individuals (P = 1.57 × 10-33). Seven genes harbored an exome-wide significant DNV burden. AC-associated genes were enriched for chromatin modifiers and converged in midgestational transcription networks essential for neural and meningeal development. Unsupervised clustering of patient phenotypes identified four AC subtypes and clinical severity correlated with the presence of a damaging DNV. These data provide insights into the coordinated regulation of brain and meningeal development and implicate epigenomic dysregulation due to DNVs in AC pathogenesis. Our results provide a preliminary indication that, in the appropriate clinical context, ACs may be considered radiographic harbingers of neurodevelopmental pathology warranting genetic testing and neurobehavioral follow-up. These data highlight the utility of a systems-level, multiomics approach to elucidate sporadic structural brain disease.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing interests.
Figures
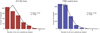

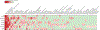



References
MAIN TEXT REFERENCES
METHODS REFERENCES
-
- Retterer K, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med 18, 696–704 (2016). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

