Evaluation of the herbicide glyphosate, (aminomethyl)phosphonic acid, and glyphosate-based formulations for genotoxic activity using in vitro assays
- PMID: 36880770
- PMCID: PMC10266336
- DOI: 10.1002/em.22534
Evaluation of the herbicide glyphosate, (aminomethyl)phosphonic acid, and glyphosate-based formulations for genotoxic activity using in vitro assays
Abstract
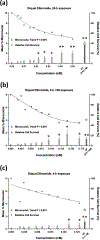
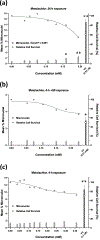
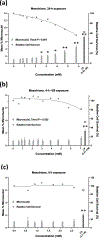
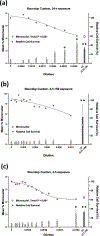
Glyphosate, the most heavily used herbicide world-wide, is applied to plants in complex formulations that promote absorption. The National Toxicology Program reported in 1992 that glyphosate, administered to rats and mice at doses up to 50,000 ppm in feed for 13 weeks, showed little evidence of toxicity, and no induction of micronuclei was observed in the mice in this study. Subsequently, mechanistic studies of glyphosate and glyphosate-based formulations (GBFs) that have focused on DNA damage and oxidative stress suggest that glyphosate may have genotoxic potential. However, few of these studies directly compared glyphosate to GBFs, or effects among GBFs. To address these data gaps, we tested glyphosate, glyphosate isopropylamine (IPA), and (aminomethyl)phosphonic acid (AMPA, a microbial metabolite of glyphosate), 9 high-use agricultural GBFs, 4 residential-use GBFs, and additional herbicides (metolachlor, mesotrione, and diquat dibromide) present in some of the GBFs in bacterial mutagenicity tests, and in human TK6 cells using a micronucleus assay and a multiplexed DNA damage assay. Our results showed no genotoxicity or notable cytotoxicity for glyphosate or AMPA at concentrations up to 10 mM, while all GBFs and herbicides other than glyphosate were cytotoxic, and some showed genotoxic activity. An in vitro to in vivo extrapolation of results for glyphosate suggests that it is of low toxicological concern for humans. In conclusion, these results demonstrate a lack of genotoxicity for glyphosate, consistent with observations in the NTP in vivo study, and suggest that toxicity associated with GBFs may be related to other components of these formulations.
Keywords: bacterial mutagenicity assay; diquat dibromide; mesotrione; metolachlor; micronucleus.
© 2023 Integrated Laboratory systems, LLC. Environmental and Molecular Mutagenesis published by Wiley Periodicals LLC on behalf of Environmental Mutagen Society. This article has been contributed to by U.S. Government employees and their work is in the public domain in the USA.
Conflict of interest statement
DECLARATION OF CONFLICTS OF INTEREST
The authors declare that they have nothing to disclose.
Figures










References
-
- Anadón A, Martínez-Larrañaga MR, Martínez MA, Castellano VJ, Martínez M, Martin MT et al. (2009) Toxicokinetics of glyphosate and its metabolite aminomethyl phosphonic acid in rats. Toxicol Lett, 190(1), 91–95. - PubMed
-
- ATSDR (Agency for Toxic Substances and Disease Registry). (2020) Toxicological profile for glyphosate. https://www.atsdr.cdc.gov/toxprofiles/tp214.pdf [accessed 10 March 2022]. - PubMed
-
- Benigni R, Bignami M, Carere A, Conti G, Conti L, Crebelli R. et al. (1979) Mutational studies with diquat and paraquat in vitro. Mutat Res, 68(3), 183–193. - PubMed
-
- Bentley R. (1990) The shikimate pathway--a metabolic tree with many branches. Crit Rev Biochem Mol Biol, 25(5), 307–384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

