Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B
- PMID: 36881464
- PMCID: PMC9991056
- DOI: 10.7554/eLife.84632
Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B
Abstract
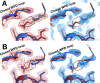
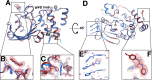
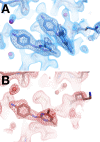
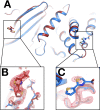
Much of our current understanding of how small-molecule ligands interact with proteins stems from X-ray crystal structures determined at cryogenic (cryo) temperature. For proteins alone, room-temperature (RT) crystallography can reveal previously hidden, biologically relevant alternate conformations. However, less is understood about how RT crystallography may impact the conformational landscapes of protein-ligand complexes. Previously, we showed that small-molecule fragments cluster in putative allosteric sites using a cryo crystallographic screen of the therapeutic target PTP1B (Keedy et al., 2018). Here, we have performed two RT crystallographic screens of PTP1B using many of the same fragments, representing the largest RT crystallographic screens of a diverse library of ligands to date, and enabling a direct interrogation of the effect of data collection temperature on protein-ligand interactions. We show that at RT, fewer ligands bind, and often more weakly - but with a variety of temperature-dependent differences, including unique binding poses, changes in solvation, new binding sites, and distinct protein allosteric conformational responses. Overall, this work suggests that the vast body of existing cryo-temperature protein-ligand structures may provide an incomplete picture, and highlights the potential of RT crystallography to help complete this picture by revealing distinct conformational modes of protein-ligand systems. Our results may inspire future use of RT crystallography to interrogate the roles of protein-ligand conformational ensembles in biological function.
Keywords: E. coli; X-ray crystallography; allostery; fragment screening; ligands; molecular biophysics; protein structure; structural biology.
© 2023, Skaist Mehlman et al.
Conflict of interest statement
TS, JB, SA, EN, SH, LD, NP, AD, RT, DA, HO, Fv, DK No competing interests declared
Figures
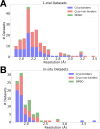









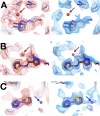

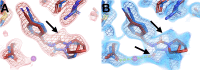




Update of
- doi: 10.1101/2022.11.02.514751
References
-
- Achdout H, Aimon A, Bar-David E, Barr H, The COVID Moonshot Consortium Open Science Discovery of Oral Non-Covalent SARS-CoV-2 Main Protease Inhibitor Therapeutics. bioRxiv. 2022 doi: 10.1101/2020.10.29.339317. - DOI
-
- Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, Millán C, Park H, Adams C, Glassman CR, DeGiovanni A, Pereira JH, Rodrigues AV, van Dijk AA, Ebrecht AC, Opperman DJ, Sagmeister T, Buhlheller C, Pavkov-Keller T, Rathinaswamy MK, Dalwadi U, Yip CK, Burke JE, Garcia KC, Grishin NV, Adams PD, Read RJ, Baker D. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

