Altered DNA repair pathway engagement by engineered CRISPR-Cas9 nucleases
- PMID: 36881621
- PMCID: PMC10242711
- DOI: 10.1073/pnas.2300605120
Altered DNA repair pathway engagement by engineered CRISPR-Cas9 nucleases
Abstract
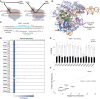
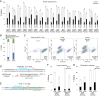
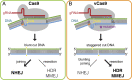
CRISPR-Cas9 introduces targeted DNA breaks that engage competing DNA repair pathways, producing a spectrum of imprecise insertion/deletion mutations (indels) and precise templated mutations (precise edits). The relative frequencies of these pathways are thought to primarily depend on genomic sequence and cell state contexts, limiting control over mutational outcomes. Here, we report that engineered Cas9 nucleases that create different DNA break structures engage competing repair pathways at dramatically altered frequencies. We accordingly designed a Cas9 variant (vCas9) that produces breaks which suppress otherwise dominant nonhomologous end-joining (NHEJ) repair. Instead, breaks created by vCas9 are predominantly repaired by pathways utilizing homologous sequences, specifically microhomology-mediated end-joining (MMEJ) and homology-directed repair (HDR). Consequently, vCas9 enables efficient precise editing through HDR or MMEJ while suppressing indels caused by NHEJ in dividing and nondividing cells. These findings establish a paradigm of targeted nucleases custom-designed for specific mutational applications.
Keywords: CRISPR; DNA repair; genome editing; molecular engineering; precise editing.
Conflict of interest statement
The authors have patent filings to disclose, the authors have filed for a patent related to this work, the authors have additional information to disclose, We note that author R.L. and reviewer D.J.S. co-authored a recent perspective article.
Figures



References
-
- Cannavo E., Cejka P., Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

