Alterations in the Proteome of Developing Neocortical Synaptosomes in the Absence of MET Signaling Revealed by Comparative Proteomics
- PMID: 36882009
- PMCID: PMC10239366
- DOI: 10.1159/000529981
Alterations in the Proteome of Developing Neocortical Synaptosomes in the Absence of MET Signaling Revealed by Comparative Proteomics
Abstract
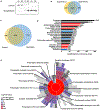
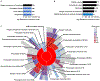
Alterations in the expression of genes encoding proteins involved in synapse formation, maturation, and function are a hallmark of many neurodevelopmental and psychiatric disorders. For example, there is reduced neocortical expression of the MET receptor tyrosine kinase (MET) transcript and protein in Autism Spectrum Disorder (ASD) and Rett syndrome. Preclinical in vivo and in vitro models manipulating MET signaling reveal that the receptor modulates excitatory synapse development and maturation in select forebrain circuits. The molecular adaptations underlying the altered synaptic development remain unknown. We performed a comparative mass spectrometry analysis of synaptosomes generated from the neocortex of wild type and Met null mice during the peak of synaptogenesis (postnatal day 14; data are available from ProteomeXchange with identifier PXD033204). The analyses revealed broad disruption of the developing synaptic proteome in the absence of MET, consistent with the localization of MET protein in pre- and postsynaptic compartments, including proteins associated with the neocortical synaptic MET interactome and those encoded by syndromic and ASD risk genes. In addition to an overrepresentation of altered proteins associated with the SNARE complex, multiple proteins in the ubiquitin-proteasome system and associated with the synaptic vesicle, as well as proteins that regulate actin filament organization and synaptic vesicle exocytosis/endocytosis, were disrupted. Taken together, the proteomic changes are consistent with structural and functional changes observed following alterations in MET signaling. We hypothesize that the molecular adaptations following Met deletion may reflect a general mechanism that produces circuit-specific molecular changes due to loss or reduction of synaptic signaling proteins.
Keywords: Developing brain with SNARE complex; Isobaric tag for relative and absolute quantitation proteomics; MET; Neocortex; Synaptosome.
© 2023 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Figures
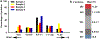


Similar articles
-
Widespread alterations in the synaptic proteome of the adolescent cerebral cortex following prenatal immune activation in rats.Brain Behav Immun. 2016 Aug;56:289-309. doi: 10.1016/j.bbi.2016.04.002. Epub 2016 Apr 4. Brain Behav Immun. 2016. PMID: 27058163
-
Hepatocyte Growth Factor Modulates MET Receptor Tyrosine Kinase and β-Catenin Functional Interactions to Enhance Synapse Formation.eNeuro. 2016 Aug 29;3(4):ENEURO.0074-16.2016. doi: 10.1523/ENEURO.0074-16.2016. eCollection 2016 Jul-Aug. eNeuro. 2016. PMID: 27595133 Free PMC article.
-
Receptor Tyrosine Kinase MET Interactome and Neurodevelopmental Disorder Partners at the Developing Synapse.Biol Psychiatry. 2016 Dec 15;80(12):933-942. doi: 10.1016/j.biopsych.2016.02.022. Epub 2016 Feb 26. Biol Psychiatry. 2016. PMID: 27086544 Free PMC article.
-
The Pleiotropic MET Receptor Network: Circuit Development and the Neural-Medical Interface of Autism.Biol Psychiatry. 2017 Mar 1;81(5):424-433. doi: 10.1016/j.biopsych.2016.08.035. Epub 2016 Sep 15. Biol Psychiatry. 2017. PMID: 27837921 Free PMC article. Review.
-
Recent Advances in Synaptosomal Proteomics in Alzheimer's Disease.Curr Protein Pept Sci. 2021;22(6):479-492. doi: 10.2174/1389203722666210618110233. Curr Protein Pept Sci. 2021. PMID: 34148536 Review.
References
-
- Qiu S, Aldinger KA, Levitt P. Modeling of autism genetic variations in mice: focusing on synaptic and microcircuit dysfunctions. Dev Neurosci. 2012;34(2–3):88–100. - PubMed
-
- Bourgeron T A synaptic trek to autism. Curr Opin Neurobiol. 2009;19(2):231–4. - PubMed
-
- Kenny EM, Cormican P, Furlong S, Heron E, Kenny G, Fahey C, et al. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol Psychiatry. 2014;19(8):872–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

