Acute stress reduces population-level metabolic and proteomic variation
- PMID: 36882728
- PMCID: PMC9993721
- DOI: 10.1186/s12859-023-05185-4
Acute stress reduces population-level metabolic and proteomic variation
Abstract
Background: Variation in omics data due to intrinsic biological stochasticity is often viewed as a challenging and undesirable feature of complex systems analyses. In fact, numerous statistical methods are utilized to minimize the variation among biological replicates.
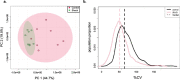
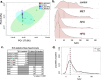
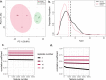
Results: We demonstrate that the common statistics relative standard deviation (RSD) and coefficient of variation (CV), which are often used for quality control or part of a larger pipeline in omics analyses, can also be used as a metric of a physiological stress response. Using an approach we term Replicate Variation Analysis (RVA), we demonstrate that acute physiological stress leads to feature-wide canalization of CV profiles of metabolomes and proteomes across biological replicates. Canalization is the repression of variation between replicates, which increases phenotypic similarity. Multiple in-house mass spectrometry omics datasets in addition to publicly available data were analyzed to assess changes in CV profiles in plants, animals, and microorganisms. In addition, proteomics data sets were evaluated utilizing RVA to identify functionality of reduced CV proteins.
Conclusions: RVA provides a foundation for understanding omics level shifts that occur in response to cellular stress. This approach to data analysis helps characterize stress response and recovery, and could be deployed to detect populations under stress, monitor health status, and conduct environmental monitoring.
Keywords: Canalization; Cellular stress; Metabolomics; Proteomics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

