Silmitasertib (CX-4945), a Clinically Used CK2-Kinase Inhibitor with Additional Effects on GSK3β and DYRK1A Kinases: A Structural Perspective
- PMID: 36883902
- PMCID: PMC10041529
- DOI: 10.1021/acs.jmedchem.2c01887
Silmitasertib (CX-4945), a Clinically Used CK2-Kinase Inhibitor with Additional Effects on GSK3β and DYRK1A Kinases: A Structural Perspective
Abstract
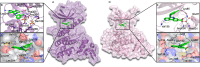
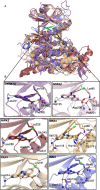
A clinical casein kinase 2 inhibitor, CX-4945 (silmitasertib), shows significant affinity toward the DYRK1A and GSK3β kinases, involved in down syndrome phenotypes, Alzheimer's disease, circadian clock regulation, and diabetes. This off-target activity offers an opportunity for studying the effect of the DYRK1A/GSK3β kinase system in disease biology and possible line extension. Motivated by the dual inhibition of these kinases, we solved and analyzed the crystal structures of DYRK1A and GSK3β with CX-4945. We built a quantum-chemistry-based model to rationalize the compound affinity for CK2α, DYRK1A, and GSK3β kinases. Our calculations identified a key element for CK2α's subnanomolar affinity to CX-4945. The methodology is expandable to other kinase selectivity modeling. We show that the inhibitor limits DYRK1A- and GSK3β-mediated cyclin D1 phosphorylation and reduces kinase-mediated NFAT signaling in the cell. Given the CX-4945's clinical and pharmacological profile, this inhibitory activity makes it an interesting candidate with potential for application in additional disease areas.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Buljan M.; Ciuffa R.; van Drogen A.; Vichalkovski A.; Mehnert M.; Rosenberger G.; Lee S.; Varjosalo M.; Pernas L. E.; Spegg V.; Snijder B.; Aebersold R.; Gstaiger M. Kinase Interaction Network Expands Functional and Disease Roles of Human Kinases. Mol. Cell 2020, 79, 504.10.1016/J.MOLCEL.2020.07.001. - DOI - PMC - PubMed
-
- Attwood M. M.; Fabbro D.; Sokolov A. V.; Knapp S.; Schiöth H. B. Trends in kinase drug discovery: targets, indications and inhibitor design. Nat. Rev. Drug Discovery 2021, 20, 839.10.1038/s41573-021-00252-y. - DOI - PubMed
- Blue Ridge Institute for Medical Research. https://brimr.org/protein-kinase-inhibitors/ (accessed Dec 28, 2022).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials

