Total and Subgenomic RNA Viral Load in Patients Infected With SARS-CoV-2 Alpha, Delta, and Omicron Variants
- PMID: 36883903
- PMCID: PMC10420395
- DOI: 10.1093/infdis/jiad061
Total and Subgenomic RNA Viral Load in Patients Infected With SARS-CoV-2 Alpha, Delta, and Omicron Variants
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomic and subgenomic RNA levels are frequently used as a correlate of infectiousness. The impact of host factors and SARS-CoV-2 lineage on RNA viral load is unclear.
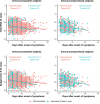
Methods: Total nucleocapsid (N) and subgenomic N (sgN) RNA levels were measured by quantitative reverse transcription polymerase chain reaction (RT-qPCR) in specimens from 3204 individuals hospitalized with coronavirus disease 2019 (COVID-19) at 21 hospitals. RT-qPCR cycle threshold (Ct) values were used to estimate RNA viral load. The impact of time of sampling, SARS-CoV-2 variant, age, comorbidities, vaccination, and immune status on N and sgN Ct values were evaluated using multiple linear regression.
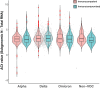
Results: Mean Ct values at presentation for N were 24.14 (SD 4.53) for non-variants of concern, 25.15 (SD 4.33) for Alpha, 25.31 (SD 4.50) for Delta, and 26.26 (SD 4.42) for Omicron. N and sgN RNA levels varied with time since symptom onset and infecting variant but not with age, comorbidity, immune status, or vaccination. When normalized to total N RNA, sgN levels were similar across all variants.
Conclusions: RNA viral loads were similar among hospitalized adults, irrespective of infecting variant and known risk factors for severe COVID-19. Total N and subgenomic RNA N viral loads were highly correlated, suggesting that subgenomic RNA measurements add little information for the purposes of estimating infectivity.
Keywords: SARS-CoV-2; subgenomic RNA; variants of concern; viral load.
© The Author(s) 2023. Published by Oxford University Press on behalf of Infectious Diseases Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Potential conflicts of interest. J. Ca. reports grants from National Institutes of Health (NIH) and Department of Defense (DoD), outside the submitted work. J. Ch. reports grants from the NIH, DoD, and Dolly Parton COVID-19 Research Fund, outside the submitted work. D. D. reports a grant from NIH, outside the submitted work. A. D. reports grants from the NIH and National Heart, Lung, and Blood Institute (NHLBI) for the ACTIV platform and PETAL network, respectively; and consulting fees for Alung Technologies, outside the submitted work. M. E. reports grants from NIH and Regeneron; personal funds for speaking at the ASPEN conference for Abbott Labs; and payment for testimony from Medical Legal Expert Witness, outside the submitted work. A. F. reports grants from NIH, outside the submitted work. M. G. reports grants from CDC, CDC-Abt Associates, and CDC-Westat; and served as cochair of the Infectious Diseases and Immunization Committee for the Texas Pediatric Society, outside the submitted work. K. G. reports grants from NIH and DoD; and support for MHSRS 2022 travel from the DoD, outside the submitted work. A. G. reports grants from NIH, DoD, AbbVie, and Faron Pharmaceuticals, outside the submitted work. M. N. G. reports grants from NHLBI, CDC, and Agency for Healthcare Research and Quality; speaking at medicine grand rounds at New York Medical College; travel support for the American Thoracic Society (ATS) executive meeting and serving as ATS Chair Critical Care Assembly; data and safety monitoring board (DSMB) membership fees from Regeneron; and participating on the scientific advisory panel for Endpoint, outside the submitted work. C. G. reports grants from NIH, CDC, AHRQ, FDA, and Campbell Alliance/Syneos Health; and consulting fees from and participating on a DSMB for Merck, outside the submitted work. D. H. reports grants from NHLBI, outside the submitted work. N. H. reports grants from Sandofi and Quidel; and a Genentech educational grant to give a lecture, outside the submitted work. A. K. reports grants from United Therapeutics, Johnson & Johnson, 4D Medical, Eli Lily, Dompe Pharmaceuticals, and GlaxoSmithKline; and serves on the guidelines committee for Chest, outside the submitted work. A. L. reports grants from CDC, National Institute of Allergy and Infectious Diseases, and Burroughs Wellcome Fund; and consulting fees from Sanofi and Roche for consulting on oseltamivir and baloxavir, respectively, outside the submitted work. E. M. reports grants from Flu Lab, Merck, and NIH outside the submitted work. T. M. reports a grant from CDC; a one-time payment for participating as a virtual webinar panelist for Clinical Updates in Heart Failure; and being a Practice Management Committee member for Society of Hospital Medicine, outside the submitted work. I. D. P. reports grants from NIH and Janssen Pharmaceuticals; and institutional support from Asahi Kasei Pharma and Regeneron, outside the submitted work. W. B. S. reports grants and support for meetings/travel from the NIH/NHLBI, outside the submitted work. J. W. reports payment for the American College of Emergency Physicians speaker honorarium and participating on the American Board of Internal Medicine Critical Care Exam Committee, outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures

References
-
- Wölfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. - PubMed

