3-Hydroxy-3-methylglutaryl coenzyme A reductase genes from Glycine max regulate plant growth and isoprenoid biosynthesis
- PMID: 36890158
- PMCID: PMC9995466
- DOI: 10.1038/s41598-023-30797-4
3-Hydroxy-3-methylglutaryl coenzyme A reductase genes from Glycine max regulate plant growth and isoprenoid biosynthesis
Abstract
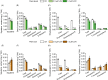
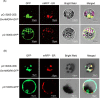
Isoprenoids, a large kind of plant natural products, are synthesized by the mevalonate (MVA) pathway in the cytoplasm and the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway in plastids. As one of the rate-limiting enzymes in the MVA pathway of soybean (Glycine max), 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) is encoded by eight isogenes (GmHMGR1-GmHMGR8). To begin, we used lovastatin (LOV), a specific inhibitor of GmHMGR, to investigate their role in soybean development. To further investigate, we overexpressed the GmHMGR4 and GmHMGR6 genes in Arabidopsis thaliana. The growth of soybean seedlings, especially the development of lateral roots, was inhibited after LOV treatment, accompanied by a decrease in sterols content and GmHMGR gene expression. After the overexpression of GmHMGR4 and GmHMGR6 in A. thaliana, the primary root length was higher than the wild type, and total sterol and squalene contents were significantly increased. In addition, we detected a significant increase in the product tocopherol from the MEP pathway. These results further support the fact that GmHMGR1-GmHMGR8 play a key role in soybean development and isoprenoid biosynthesis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

