Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy
- PMID: 36890231
- PMCID: PMC10258627
- DOI: 10.1038/s41586-023-05788-0
Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy
Erratum in
-
Author Correction: Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy.Nature. 2024 Oct;634(8036):E15. doi: 10.1038/s41586-024-08200-7. Nature. 2024. PMID: 39415023 No abstract available.
Abstract
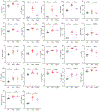
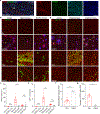
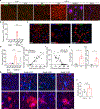
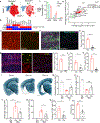
Extracellular deposition of amyloid-β as neuritic plaques and intracellular accumulation of hyperphosphorylated, aggregated tau as neurofibrillary tangles are two of the characteristic hallmarks of Alzheimer's disease1,2. The regional progression of brain atrophy in Alzheimer's disease highly correlates with tau accumulation but not amyloid deposition3-5, and the mechanisms of tau-mediated neurodegeneration remain elusive. Innate immune responses represent a common pathway for the initiation and progression of some neurodegenerative diseases. So far, little is known about the extent or role of the adaptive immune response and its interaction with the innate immune response in the presence of amyloid-β or tau pathology6. Here we systematically compared the immunological milieux in the brain of mice with amyloid deposition or tau aggregation and neurodegeneration. We found that mice with tauopathy but not those with amyloid deposition developed a unique innate and adaptive immune response and that depletion of microglia or T cells blocked tau-mediated neurodegeneration. Numbers of T cells, especially those of cytotoxic T cells, were markedly increased in areas with tau pathology in mice with tauopathy and in the Alzheimer's disease brain. T cell numbers correlated with the extent of neuronal loss, and the cells dynamically transformed their cellular characteristics from activated to exhausted states along with unique TCR clonal expansion. Inhibition of interferon-γ and PDCD1 signalling both significantly ameliorated brain atrophy. Our results thus reveal a tauopathy- and neurodegeneration-related immune hub involving activated microglia and T cell responses, which could serve as therapeutic targets for preventing neurodegeneration in Alzheimer's disease and primary tauopathies.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Conflicts of Interest
D.M.H. is as an inventor on a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies. D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics. D.M.H. is on the scientific advisory board of Denali and Cajal Neuroscience and consults for Genentech and Alector. J.K. is a member of a scientific advisory group for PureTech. All other authors declare no competing interests.
Figures











Comment in
-
Activated immune cells drive neurodegeneration in an Alzheimer's model.Nature. 2023 Mar;615(7953):588-589. doi: 10.1038/d41586-023-00600-5. Nature. 2023. PMID: 36890310 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

