Immunolyser: A web-based computational pipeline for analysing and mining immunopeptidomic data
- PMID: 36890882
- PMCID: PMC9988424
- DOI: 10.1016/j.csbj.2023.02.033
Immunolyser: A web-based computational pipeline for analysing and mining immunopeptidomic data
Abstract
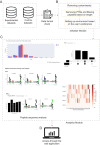
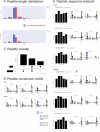
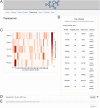
Immunopeptidomics has made tremendous contributions to our understanding of antigen processing and presentation, by identifying and quantifying antigenic peptides presented on the cell surface by Major Histocompatibility Complex (MHC) molecules. Large and complex immunopeptidomics datasets can now be routinely generated using Liquid Chromatography-Mass Spectrometry techniques. The analysis of this data - often consisting of multiple replicates/conditions - rarely follows a standard data processing pipeline, hindering the reproducibility and depth of analysis of immunopeptidomic data. Here, we present Immunolyser, an automated pipeline designed to facilitate computational analysis of immunopeptidomic data with a minimal initial setup. Immunolyser brings together routine analyses, including peptide length distribution, peptide motif analysis, sequence clustering, peptide-MHC binding affinity prediction, and source protein analysis. Immunolyser provides a user-friendly and interactive interface via its webserver and is freely available for academic purposes at https://immunolyser.erc.monash.edu/. The open-access source code can be downloaded at our GitHub repository: https://github.com/prmunday/Immunolyser. We anticipate that Immunolyser will serve as a prominent computational pipeline to facilitate effortless and reproducible analysis of immunopeptidomic data.
Keywords: Antigen processing and presentation; Immunopeptidomics; Peptide analysis.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Liepe J., Sidney J., Lorenz F.K., et al. Mapping the MHC class I–spliced immunopeptidome of cancer cells. Cancer Immunol Res. 2019;7:62–76. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
