Inoculum production of Phytophthora medicaginis can be used to screen for partial resistance in chickpea genotypes
- PMID: 36890901
- PMCID: PMC9986325
- DOI: 10.3389/fpls.2023.1115417
Inoculum production of Phytophthora medicaginis can be used to screen for partial resistance in chickpea genotypes
Abstract
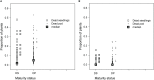
Phytophthora root rot caused by Phytophthora medicaginis is an important disease of chickpeas (Cicer arietinum) in Australia with limited management options, increasing reliance on breeding for improved levels of genetic resistance. Resistance based on chickpea-Cicer echinospermum crosses is partial with a quantitative genetic basis provided by C. echinospermum and some disease tolerance traits originating from C. arietinum germplasm. Partial resistance is hypothesised to reduce pathogen proliferation, while tolerant germplasm may contribute some fitness traits, such as an ability to maintain yield despite pathogen proliferation. To test these hypotheses, we used P. medicaginis DNA concentrations in the soil as a parameter for pathogen proliferation and disease assessments on lines of two recombinant inbred populations of chickpea-C. echinospermum crosses to compare the reactions of selected recombinant inbred lines and parents. Our results showed reduced inoculum production in a C. echinospermum backcross parent relative to the C. arietinum variety Yorker. Recombinant inbred lines with consistently low levels of foliage symptoms had significantly lower levels of soil inoculum compared to lines with high levels of visible foliage symptoms. In a separate experiment, a set of superior recombinant inbred lines with consistently low levels of foliage symptoms was tested for soil inoculum reactions relative to control normalised yield loss. The in-crop P. medicaginis soil inoculum concentrations across genotypes were significantly and positively related to yield loss, indicating a partial resistance-tolerance spectrum. Disease incidence and the rankings for in-crop soil inoculum were correlated strongly to yield loss. These results indicate that soil inoculum reactions may be useful to identify genotypes with high levels of partial resistance.
Keywords: pathogen proliferation; phenotyping; quantitative resistance; root disease; tolerance.
Copyright © 2023 Bithell, Drenth, Backhouse, Harden and Hobson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Ali M. A., Nawab N. N., Abbas A., Zulkiffal M., Sajjad M. (2009). Evaluation of selection criteria in Cicer arietinum l. using correlation coefficients and path analysis. Aust. J. Crop Sci. 3 (2), 65–70.
-
- Amalraj A., Taylor J., Bithell S., Li Y., Moore K., Hobson K., et al. (2019). Mapping resistance to phytophthora root rot identifies independent loci from cultivated (Cicer arietinum l.) and wild (Cicer echinospermum P.H. Davis) chickpea. Theor. Appl. Genet. 132 (4), 1017–1033. doi: 10.1007/s00122-018-3256-6 - DOI - PubMed
-
- Anon (2018). “GenStat committee,” in The guide to GenStat release 19.1 (Oxford: VSN International; ).
-
- Bithell S. L., Backhouse D., Harden S., Drenth A., Moore K., Flavel R. J., et al. (2022). Aggressiveness of Phytophthora medicaginis on chickpea: Phenotyping method determines isolate ranking and host genotype-isolate interactions. Plant Pathol. 71, 1076–1091. doi: 10.1111/ppa.13547 - DOI
-
- Bithell S. L., Moore K., McKay A., Harden S., Simpfendorfer S. (2021). Phytophthora root rot of chickpea: Inoculum concentration and seasonally dependent success for qPCR based predictions of disease and yield loss. Australas. Plant Pathol. 50 (1), 91–103. doi: 10.1007/s13313-020-00752-2 - DOI
LinkOut - more resources
Full Text Sources

