Exploiting the mediating role of the metabolome to unravel transcript-to-phenotype associations
- PMID: 36891970
- PMCID: PMC9998083
- DOI: 10.7554/eLife.81097
Exploiting the mediating role of the metabolome to unravel transcript-to-phenotype associations
Abstract
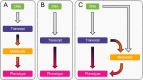
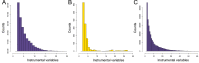
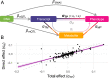
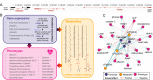
Despite the success of genome-wide association studies (GWASs) in identifying genetic variants associated with complex traits, understanding the mechanisms behind these statistical associations remains challenging. Several methods that integrate methylation, gene expression, and protein quantitative trait loci (QTLs) with GWAS data to determine their causal role in the path from genotype to phenotype have been proposed. Here, we developed and applied a multi-omics Mendelian randomization (MR) framework to study how metabolites mediate the effect of gene expression on complex traits. We identified 216 transcript-metabolite-trait causal triplets involving 26 medically relevant phenotypes. Among these associations, 58% were missed by classical transcriptome-wide MR, which only uses gene expression and GWAS data. This allowed the identification of biologically relevant pathways, such as between ANKH and calcium levels mediated by citrate levels and SLC6A12 and serum creatinine through modulation of the levels of the renal osmolyte betaine. We show that the signals missed by transcriptome-wide MR are found, thanks to the increase in power conferred by integrating multiple omics layer. Simulation analyses show that with larger molecular QTL studies and in case of mediated effects, our multi-omics MR framework outperforms classical MR approaches designed to detect causal relationships between single molecular traits and complex phenotypes.
Keywords: Mediation; Mendelian Randomization; gene expression; genetics; genomics; human; metabolomics.
© 2023, Auwerx et al.
Conflict of interest statement
CA, MS, TW, AR, ZK, EP No competing interests declared
Figures

Comment in
-
The next step in Mendelian randomization.Elife. 2023 Mar 9;12:e86416. doi: 10.7554/eLife.86416. Elife. 2023. PMID: 36891986 Free PMC article.
References
-
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

