SARS-CoV-2 Exposure in Norway Rats (Rattus norvegicus) from New York City
- PMID: 36892291
- PMCID: PMC10127689
- DOI: 10.1128/mbio.03621-22
SARS-CoV-2 Exposure in Norway Rats (Rattus norvegicus) from New York City
Abstract
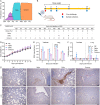
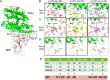
Millions of Norway rats (Rattus norvegicus) inhabit New York City (NYC), presenting the potential for transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from humans to rats. We evaluated SARS-CoV-2 exposure among 79 rats captured from NYC during the fall of 2021. Our results showed that 13 of the 79 rats (16.5%) tested IgG- or IgM-positive, and partial SARS-CoV-2 genomes were recovered from all 4 rats that were qRT-PCR (reverse transcription-quantitative PCR)-positive. Genomic analyses suggest these viruses were associated with genetic lineage B, which was predominant in NYC in the spring of 2020 during the early pandemic period. To further investigate rat susceptibility to SARS-CoV-2 variants, we conducted a virus challenge study and showed that Alpha, Delta, and Omicron variants can cause infections in wild-type Sprague Dawley (SD) rats, including high replication levels in the upper and lower respiratory tracts and induction of both innate and adaptive immune responses. Additionally, the Delta variant resulted in the highest infectivity. In summary, our results indicate that rats are susceptible to infection with Alpha, Delta, and Omicron variants, and wild Norway rats in the NYC municipal sewer systems have been exposed to SARS-CoV-2. Our findings highlight the need for further monitoring of SARS-CoV-2 in urban rat populations and for evaluating the potential risk of secondary zoonotic transmission from these rat populations back to humans. IMPORTANCE The host tropism expansion of SARS-CoV-2 raises concern for the potential risk of reverse-zoonotic transmission of emerging variants into rodent species, including wild rat species. In this study, we present both genetic and serological evidence for SARS-CoV-2 exposure to the New York City wild rat population, and these viruses may be linked to the viruses that were circulating during the early stages of the pandemic. We also demonstrated that rats are susceptible to additional variants (i.e., Alpha, Delta, and Omicron) that have been predominant in humans and that susceptibility to infection varies by variant. Our findings highlight the reverse zoonosis of SARS-CoV-2 to urban rats and the need for further monitoring of SARS-CoV-2 in rat populations for potential secondary zoonotic transmission to humans.
Keywords: Delta; Norway rats; Omicron; Rattus norvegicus; SARS-CoV-2; brown rats; rat COVID-19; rat coronavirus; reverse zoonosis; surveillance; wildlife.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Update of
-
SARS-CoV-2 exposure in Norway rats (Rattus norvegicus) from New York City.bioRxiv [Preprint]. 2022 Dec 10:2022.11.18.517156. doi: 10.1101/2022.11.18.517156. bioRxiv. 2022. Update in: mBio. 2023 Apr 25;14(2):e0362122. doi: 10.1128/mbio.03621-22. PMID: 36451891 Free PMC article. Updated. Preprint.
References
-
- Johns Hopkins University of Medicine. 2022. Coronavirus resource center. Available from https://coronavirus.jhu.edu/data. Accessed 10 October 2022. Johns Hopkins University, Baltimore, MD.
-
- Chandler JC, Bevins SN, Ellis JW, Linder TJ, Tell RM, Jenkins-Moore M, Root JJ, Lenoch JB, Robbe-Austerman S, DeLiberto TJ, Gidlewski T, Kim TM, Shriner SA. 2021. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc Natl Acad Sci USA 118:e2114828118. doi: 10.1073/pnas.2114828118. - DOI - PMC - PubMed
-
- Hale VL, Dennis PM, McBride DS, Nolting JM, Madden C, Huey D, Ehrlich M, Grieser J, Winston J, Lombardi D, Gibson S, Saif L, Killian ML, Lantz K, Tell RM, Torchetti M, Robbe-Austerman S, Nelson MI, Faith SA, Bowman AS. 2022. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 602:481–486. doi: 10.1038/s41586-021-04353-x. - DOI - PMC - PubMed
-
- Hammer AS, Quaade ML, Rasmussen TB, Fonager J, Rasmussen M, Mundbjerg K, Lohse L, Strandbygaard B, Jorgensen CS, Alfaro-Nunez A, Rosenstierne MW, Boklund A, Halasa T, Fomsgaard A, Belsham GJ, Botner A. 2021. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerg Infect Dis 27:547–551. doi: 10.3201/eid2702.203794. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
