A disease-associated XPA allele interferes with TFIIH binding and primarily affects transcription-coupled nucleotide excision repair
- PMID: 36893274
- PMCID: PMC10089173
- DOI: 10.1073/pnas.2208860120
A disease-associated XPA allele interferes with TFIIH binding and primarily affects transcription-coupled nucleotide excision repair
Abstract
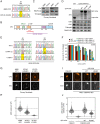
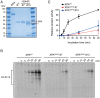
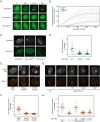
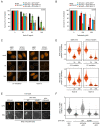
XPA is a central scaffold protein that coordinates the assembly of repair complexes in the global genome (GG-NER) and transcription-coupled nucleotide excision repair (TC-NER) subpathways. Inactivating mutations in XPA cause xeroderma pigmentosum (XP), which is characterized by extreme UV sensitivity and a highly elevated skin cancer risk. Here, we describe two Dutch siblings in their late forties carrying a homozygous H244R substitution in the C-terminus of XPA. They present with mild cutaneous manifestations of XP without skin cancer but suffer from marked neurological features, including cerebellar ataxia. We show that the mutant XPA protein has a severely weakened interaction with the transcription factor IIH (TFIIH) complex leading to an impaired association of the mutant XPA and the downstream endonuclease ERCC1-XPF with NER complexes. Despite these defects, the patient-derived fibroblasts and reconstituted knockout cells carrying the XPA-H244R substitution show intermediate UV sensitivity and considerable levels of residual GG-NER (~50%), in line with the intrinsic properties and activities of the purified protein. By contrast, XPA-H244R cells are exquisitely sensitive to transcription-blocking DNA damage, show no detectable recovery of transcription after UV irradiation, and display a severe deficiency in TC-NER-associated unscheduled DNA synthesis. Our characterization of a new case of XPA deficiency that interferes with TFIIH binding and primarily affects the transcription-coupled subpathway of nucleotide excision repair, provides an explanation of the dominant neurological features in these patients, and reveals a specific role for the C-terminus of XPA in TC-NER.
Keywords: DNA repair; nucleotide excision repair; transcription factor II H; transcription-coupled repair; xeroderma pigmentosum protein A.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy.Hum Mutat. 2008 Oct;29(10):1194-208. doi: 10.1002/humu.20768. Hum Mutat. 2008. PMID: 18470933 Free PMC article.
-
The role of Transcription Factor IIH complex in nucleotide excision repair.Environ Mol Mutagen. 2024 Apr;65 Suppl 1(Suppl 1):72-81. doi: 10.1002/em.22568. Epub 2023 Aug 23. Environ Mol Mutagen. 2024. PMID: 37545038 Free PMC article. Review.
-
Influence of XPB helicase on recruitment and redistribution of nucleotide excision repair proteins at sites of UV-induced DNA damage.DNA Repair (Amst). 2007 Sep 1;6(9):1359-70. doi: 10.1016/j.dnarep.2007.03.025. Epub 2007 May 16. DNA Repair (Amst). 2007. PMID: 17509950 Free PMC article.
-
Mouse model for the DNA repair/basal transcription disorder trichothiodystrophy reveals cancer predisposition.Cancer Res. 1999 Jul 15;59(14):3489-94. Cancer Res. 1999. PMID: 10416615
-
Common pathways for ultraviolet skin carcinogenesis in the repair and replication defective groups of xeroderma pigmentosum.J Dermatol Sci. 2000 May;23(1):1-11. doi: 10.1016/s0923-1811(99)00088-2. J Dermatol Sci. 2000. PMID: 10699759 Review.
Cited by
-
Case report: Xeroderma pigmentosum Group A with erythropoietic protoporphyria in a young Chinese patient.Front Endocrinol (Lausanne). 2024 Jul 26;15:1418254. doi: 10.3389/fendo.2024.1418254. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39129919 Free PMC article.
-
Cancer-associated fibroblast derived CXCL14 drives cisplatin chemoresistance by enhancing nucleotide excision repair in bladder cancer.J Exp Clin Cancer Res. 2025 Sep 2;44(1):265. doi: 10.1186/s13046-025-03487-4. J Exp Clin Cancer Res. 2025. PMID: 40898244 Free PMC article.
-
Cross-species investigation into the requirement of XPA for nucleotide excision repair.Nucleic Acids Res. 2024 Jan 25;52(2):677-689. doi: 10.1093/nar/gkad1104. Nucleic Acids Res. 2024. PMID: 37994737 Free PMC article.
-
STK19 facilitates the clearance of lesion-stalled RNAPII during transcription-coupled DNA repair.Cell. 2024 Dec 12;187(25):7107-7125.e25. doi: 10.1016/j.cell.2024.10.018. Epub 2024 Nov 14. Cell. 2024. PMID: 39547229 Free PMC article.
-
Protein-protein interactions in the core nucleotide excision repair pathway.DNA Repair (Amst). 2024 Sep;141:103728. doi: 10.1016/j.dnarep.2024.103728. Epub 2024 Jul 14. DNA Repair (Amst). 2024. PMID: 39029374 Free PMC article. Review.
References
-
- Sugasawa K., et al. , UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121, 387–400 (2005). - PubMed
-
- Volker M., et al. , Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8, 213–224 (2001). - PubMed
-
- Sugasawa K., et al. , Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2, 223–232 (1998). - PubMed
-
- Mellon I., Spivak G., Hanawalt P. C., Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249 (1987). - PubMed
-
- Nakazawa Y., et al. , Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat. Genet. 44, 586–592 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

