Molecular basis of polycomb group protein-mediated fetal hemoglobin repression
- PMID: 36893455
- PMCID: PMC10273169
- DOI: 10.1182/blood.2022019578
Molecular basis of polycomb group protein-mediated fetal hemoglobin repression
Abstract
The switch from fetal hemoglobin (HbF) to adult hemoglobin (HbA) is a paradigm for developmental gene expression control with relevance to sickle cell disease and β-thalassemia. Polycomb repressive complex (PRC) proteins regulate this switch, and an inhibitor of PRC2 has entered a clinical trial for HbF activation. Yet, how PRC complexes function in this process, their target genes, and relevant subunit composition are unknown. Here, we identified the PRC1 subunit BMI1 as a novel HbF repressor. We uncovered the RNA binding proteins LIN28B, IGF2BP1, and IGF2BP3 genes as direct BMI1 targets, and demonstrate that they account for the entirety of BMI1's effect on HbF regulation. BMI1 functions as part of the canonical PRC1 (cPRC1) subcomplex as revealed by the physical and functional dissection of BMI1 protein partners. Lastly, we demonstrate that BMI1/cPRC1 acts in concert with PRC2 to repress HbF through the same target genes. Our study illuminates how PRC silences HbF, highlighting an epigenetic mechanism involved in hemoglobin switching.
© 2023 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: G.A.B. received funding from Pfizer and Fulcrum Therapeutics. The remaining authors declare no competing financial interests.
Figures
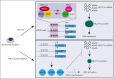
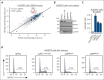
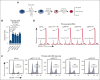
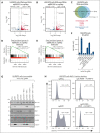

Comment in
-
Help on the way to unsilence HbF.Blood. 2023 Jun 1;141(22):2670-2672. doi: 10.1182/blood.2023020345. Blood. 2023. PMID: 37530647 Free PMC article. No abstract available.
References
-
- Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian β-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205(1-2):73–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

