Circadian clock molecule REV-ERBα regulates lung fibrotic progression through collagen stabilization
- PMID: 36894533
- PMCID: PMC9996598
- DOI: 10.1038/s41467-023-36896-0
Circadian clock molecule REV-ERBα regulates lung fibrotic progression through collagen stabilization
Abstract
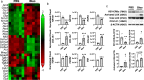
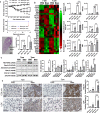
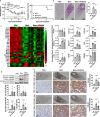
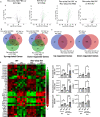
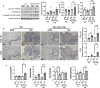
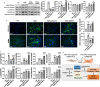
Molecular clock REV-ERBα is central to regulating lung injuries, and decreased REV-ERBα abundance mediates sensitivity to pro-fibrotic insults and exacerbates fibrotic progression. In this study, we determine the role of REV-ERBα in fibrogenesis induced by bleomycin and Influenza A virus (IAV). Bleomycin exposure decreases the abundance of REV-ERBα, and mice dosed with bleomycin at night display exacerbated lung fibrogenesis. Rev-erbα agonist (SR9009) treatment prevents bleomycin induced collagen overexpression in mice. Rev-erbα global heterozygous (Rev-erbα Het) mice infected with IAV showed augmented levels of collagens and lysyl oxidases compared with WT-infected mice. Furthermore, Rev-erbα agonist (GSK4112) prevents collagen and lysyl oxidase overexpression induced by TGFβ in human lung fibroblasts, whereas the Rev-erbα antagonist exacerbates it. Overall, these results indicate that loss of REV-ERBα exacerbates the fibrotic responses by promoting collagen and lysyl oxidase expression, whereas Rev-erbα agonist prevents it. This study provides the potential of Rev-erbα agonists in the treatment of pulmonary fibrosis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

